Network analysis identifies protein clusters of functional importance in juvenile idiopathic arthritis
- PMID: 24886659
- PMCID: PMC4062926
- DOI: 10.1186/ar4559
Network analysis identifies protein clusters of functional importance in juvenile idiopathic arthritis
Abstract
Introduction: Our objective was to utilise network analysis to identify protein clusters of greatest potential functional relevance in the pathogenesis of oligoarticular and rheumatoid factor negative (RF-ve) polyarticular juvenile idiopathic arthritis (JIA).
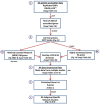
Methods: JIA genetic association data were used to build an interactome network model in BioGRID 3.2.99. The top 10% of this protein:protein JIA Interactome was used to generate a minimal essential network (MEN). Reactome FI Cytoscape 2.83 Plugin and the Disease Association Protein-Protein Link Evaluator (Dapple) algorithm were used to assess the functionality of the biological pathways within the MEN and to statistically rank the proteins. JIA gene expression data were integrated with the MEN and clusters of functionally important proteins derived using MCODE.
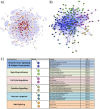
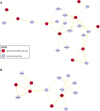
Results: A JIA interactome of 2,479 proteins was built from 348 JIA associated genes. The MEN, representing the most functionally related components of the network, comprised of seven clusters, with distinct functional characteristics. Four gene expression datasets from peripheral blood mononuclear cells (PBMC), neutrophils and synovial fluid monocytes, were mapped onto the MEN and a list of genes enriched for functional significance identified. This analysis revealed the genes of greatest potential functional importance to be PTPN2 and STAT1 for oligoarticular JIA and KSR1 for RF-ve polyarticular JIA. Clusters of 23 and 14 related proteins were derived for oligoarticular and RF-ve polyarticular JIA respectively.
Conclusions: This first report of the application of network biology to JIA, integrating genetic association findings and gene expression data, has prioritised protein clusters for functional validation and identified new pathways for targeted pharmacological intervention.
Figures

Similar articles
-
Prior to extension, Transcriptomes of fibroblast-like Synoviocytes from extended and Polyarticular juvenile idiopathic arthritis are indistinguishable.Pediatr Rheumatol Online J. 2018 Jan 8;16(1):3. doi: 10.1186/s12969-017-0217-6. Pediatr Rheumatol Online J. 2018. PMID: 29310668 Free PMC article.
-
Biologic similarities based on age at onset in oligoarticular and polyarticular subtypes of juvenile idiopathic arthritis.Arthritis Rheum. 2010 Nov;62(11):3249-58. doi: 10.1002/art.27657. Arthritis Rheum. 2010. PMID: 20662067 Free PMC article.
-
Using Chromatin Architecture to Understand the Genetics and Transcriptomics of Juvenile Idiopathic Arthritis.Front Immunol. 2018 Dec 14;9:2964. doi: 10.3389/fimmu.2018.02964. eCollection 2018. Front Immunol. 2018. PMID: 30619322 Free PMC article. Review.
-
Network analysis and juvenile idiopathic arthritis (JIA): a new horizon for the understanding of disease pathogenesis and therapeutic target identification.Pediatr Rheumatol Online J. 2016 Jul 2;14(1):40. doi: 10.1186/s12969-016-0078-4. Pediatr Rheumatol Online J. 2016. PMID: 27411317 Free PMC article. Review.
-
Brief Report: The Genetic Profile of Rheumatoid Factor-Positive Polyarticular Juvenile Idiopathic Arthritis Resembles That of Adult Rheumatoid Arthritis.Arthritis Rheumatol. 2018 Jun;70(6):957-962. doi: 10.1002/art.40443. Epub 2018 Apr 21. Arthritis Rheumatol. 2018. PMID: 29426059 Free PMC article.
Cited by
-
Integrated genomics identifies convergence of ankylosing spondylitis with global immune mediated disease pathways.Sci Rep. 2015 May 18;5:10314. doi: 10.1038/srep10314. Sci Rep. 2015. PMID: 25980808 Free PMC article.
-
Adhesion, proliferation, and apoptosis in different molecular portraits of breast cancer treated with silver nanoparticles and its pathway-network analysis.Int J Nanomedicine. 2018 Feb 22;13:1081-1095. doi: 10.2147/IJN.S152237. eCollection 2018. Int J Nanomedicine. 2018. PMID: 29503542 Free PMC article.
-
Transcriptional profiles of JIA patient blood with subsequent poor response to methotrexate.Rheumatology (Oxford). 2017 Sep 1;56(9):1542-1551. doi: 10.1093/rheumatology/kex206. Rheumatology (Oxford). 2017. PMID: 28582527 Free PMC article.
-
Complexity and Specificity of the Neutrophil Transcriptomes in Juvenile Idiopathic Arthritis.Sci Rep. 2016 Jun 7;6:27453. doi: 10.1038/srep27453. Sci Rep. 2016. PMID: 27271962 Free PMC article.
-
Integrative omics analysis of rheumatoid arthritis identifies non-obvious therapeutic targets.PLoS One. 2015 Apr 22;10(4):e0124254. doi: 10.1371/journal.pone.0124254. eCollection 2015. PLoS One. 2015. PMID: 25901943 Free PMC article.
References
-
- Petty RE, Southwood TR, Manners P, Baum J, Glass DN, Goldenberg J, He X, Maldonado-Cocco J, Orozco-Alcala J, Prieur AM, Suarez-Almazor ME, Woo P. International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J Rheumatol. 2004;31:390–392. - PubMed
-
- Hyrich KL, Lal SD, Foster HE, Thornton J, Adib N, Baildam E, Gardner-Medwin J, Wedderburn LR, Chieng A, Davidson J, Thomson W. Disease activity and disability in children with juvenile idiopathic arthritis one year following presentation to paediatric rheumatology. Results from the Childhood Arthritis Prospective Study. Rheumatology (Oxford) 2010;49:116–122. doi: 10.1093/rheumatology/kep352. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

