Lysine-specific demethylase 1-mediated demethylation of histone H3 lysine 9 contributes to interleukin 1β-induced microsomal prostaglandin E synthase 1 expression in human osteoarthritic chondrocytes
- PMID: 24886859
- PMCID: PMC4060543
- DOI: 10.1186/ar4564
Lysine-specific demethylase 1-mediated demethylation of histone H3 lysine 9 contributes to interleukin 1β-induced microsomal prostaglandin E synthase 1 expression in human osteoarthritic chondrocytes
Abstract
Introduction: Microsomal prostaglandin E synthase 1 (mPGES-1) catalyzes the terminal step in the biosynthesis of PGE2, a critical mediator in the pathophysiology of osteoarthritis (OA). Histone methylation plays an important role in epigenetic gene regulation. In this study, we investigated the roles of histone H3 lysine 9 (H3K9) methylation in interleukin 1β (IL-1β)-induced mPGES-1 expression in human chondrocytes.
Methods: Chondrocytes were stimulated with IL-1β, and the expression of mPGES-1 mRNA was evaluated using real-time RT-PCR. H3K9 methylation and the recruitment of the histone demethylase lysine-specific demethylase 1 (LSD1) to the mPGES-1 promoter were evaluated using chromatin immunoprecipitation assays. The role of LSD1 was further evaluated using the pharmacological inhibitors tranylcypromine and pargyline and small interfering RNA (siRNA)-mediated gene silencing. The LSD1 level in cartilage was determined by RT-PCR and immunohistochemistry.
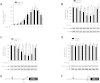
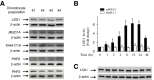
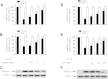
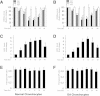
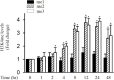
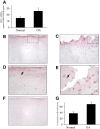
Results: The induction of mPGES-1 expression by IL-1β correlated with decreased levels of mono- and dimethylated H3K9 at the mPGES-1 promoter. These changes were concomitant with the recruitment of the histone demethylase LSD1. Treatment with tranylcypromine and pargyline, which are potent inhibitors of LSD1, prevented IL-1β-induced H3K9 demethylation at the mPGES-1 promoter and expression of mPGES-1. Consistently, LSD1 gene silencing with siRNA prevented IL-1β-induced H3K9 demethylation and mPGES-1 expression, suggesting that LSD1 mediates IL-1β-induced mPGES-1 expression via H3K9 demethylation. We show that the level of LSD1 was elevated in OA compared to normal cartilage.
Conclusion: These results indicate that H3K9 demethylation by LSD1 contributes to IL-1β-induced mPGES-1 expression and suggest that this pathway could be a potential target for pharmacological intervention in the treatment of OA and possibly other arthritic conditions.
Figures

Similar articles
-
Valproic acid suppresses interleukin-1ß-induced microsomal prostaglandin E2 synthase-1 expression in chondrocytes through upregulation of NAB1.J Rheumatol. 2011 Mar;38(3):492-502. doi: 10.3899/jrheum.100907. Epub 2011 Jan 15. J Rheumatol. 2011. PMID: 21239760
-
Expression and regulation of microsomal prostaglandin E synthase-1 in human osteoarthritic cartilage and chondrocytes.J Rheumatol. 2005 May;32(5):887-95. J Rheumatol. 2005. PMID: 15868626
-
HDAC4 contributes to IL-1-induced mPGES-1 expression in human synovial fibroblasts through up-regulation of Egr-1 transcriptional activity.J Cell Biochem. 2009 Feb 15;106(3):453-63. doi: 10.1002/jcb.22027. J Cell Biochem. 2009. PMID: 19115247
-
LSD1: more than demethylation of histone lysine residues.Exp Mol Med. 2020 Dec;52(12):1936-1947. doi: 10.1038/s12276-020-00542-2. Epub 2020 Dec 14. Exp Mol Med. 2020. PMID: 33318631 Free PMC article. Review.
-
Post-translational modifications of lysine-specific demethylase 1.Biochim Biophys Acta Gene Regul Mech. 2023 Dec;1866(4):194968. doi: 10.1016/j.bbagrm.2023.194968. Epub 2023 Aug 10. Biochim Biophys Acta Gene Regul Mech. 2023. PMID: 37572976 Review.
Cited by
-
The epigenetic players and the chromatin marks involved in the articular cartilage during osteoarthritis.Front Physiol. 2023 Jan 17;14:1070241. doi: 10.3389/fphys.2023.1070241. eCollection 2023. Front Physiol. 2023. PMID: 36733912 Free PMC article. Review.
-
Echinocystic Acid Inhibits IL-1β-Induced COX-2 and iNOS Expression in Human Osteoarthritis Chondrocytes.Inflammation. 2016 Apr;39(2):543-9. doi: 10.1007/s10753-015-0278-y. Inflammation. 2016. PMID: 26499345
-
Current understanding of osteoarthritis pathogenesis and relevant new approaches.Bone Res. 2022 Sep 20;10(1):60. doi: 10.1038/s41413-022-00226-9. Bone Res. 2022. PMID: 36127328 Free PMC article. Review.
-
Histone Modifications and Chondrocyte Fate: Regulation and Therapeutic Implications.Front Cell Dev Biol. 2021 Apr 16;9:626708. doi: 10.3389/fcell.2021.626708. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 33937229 Free PMC article. Review.
-
The Lysine Specific Demethylase-1 Negatively Regulates the COL9A1 Gene in Human Articular Chondrocytes.Int J Mol Sci. 2020 Aug 31;21(17):6322. doi: 10.3390/ijms21176322. Int J Mol Sci. 2020. PMID: 32878268 Free PMC article.
References
-
- Lawrence RC, Helmick CG, Arnett FC, Deyo RA, Felson DT, Giannini EH, Heyse SP, Hirsch R, Hochberg MC, Hunder GG, Hunder GG, Liang MH, Pillemer SR, Steen VD, Wolfe F. Estimates of the prevalence of arthritis and selected musculoskeletal disorders in the United States. Arthritis Rheum. 1998;41:778–799. doi: 10.1002/1529-0131(199805)41:5<778::AID-ART4>3.0.CO;2-V. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

