Resequencing the susceptibility gene, ITGAM, identifies two functionally deleterious rare variants in systemic lupus erythematosus cases
- PMID: 24886912
- PMCID: PMC4060450
- DOI: 10.1186/ar4566
Resequencing the susceptibility gene, ITGAM, identifies two functionally deleterious rare variants in systemic lupus erythematosus cases
Abstract
Introduction: The majority of the genetic variance of systemic lupus erythematosus (SLE) remains unexplained by the common disease-common variant hypothesis. Rare variants, which are not detectable by genome-wide association studies because of their low frequencies, are predicted to explain part of this "missing heritability." However, recent studies identifying rare variants within known disease-susceptibility loci have failed to show genetic associations because of their extremely low frequencies, leading to the questioning of the contribution of rare variants to disease susceptibility. A common (minor allele frequency = 17.4% in cases) nonsynonymous coding variant rs1143679 (R77H) in ITGAM (CD11b), which forms half of the heterodimeric integrin receptor, complement receptor 3 (CR3), is robustly associated with SLE and has been shown to impair CR3-mediated phagocytosis.
Methods: We resequenced ITGAM in 73 SLE cases and identified two previously unidentified, case-specific nonsynonymous variants, F941V and G1145S. Both variants were genotyped in 2,107 and 949 additional SLE cases, respectively, to estimate their frequencies in a disease population. An in vitro model was used to assess the impact of F941V and G1145S, together with two nonsynonymous ITGAM polymorphisms, A858V (rs1143683) and M441T (rs11861251), on CR3-mediated phagocytosis. A paired two-tailed t test was used to compare the phagocytic capabilities of each variant with that of wild-type CR3.
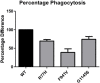
Results: Both rare variants, F941V and G1145S, significantly impair CR3-mediated phagocytosis in an in vitro model (61% reduction, P = 0.006; 26% reduction, P = 0.0232). However, neither of the common variants, M441T and A858V, had an effect on phagocytosis. Neither rare variant was observed again in the genotyping of additional SLE cases, suggesting that their frequencies are extremely low.
Conclusions: Our results add further evidence to the functional importance of ITGAM in SLE pathogenesis through impaired phagocytosis. Additionally, this study provides a new example of the identification of rare variants in common-allele-associated loci, which, because of their extremely low frequencies, are not statistically associated. However, the demonstration of their functional effects adds support to their contribution to disease risk, and questions the current notion of dismissing the contribution of very rare variants on purely statistical analyses.
Figures




Similar articles
-
Multiple lupus-associated ITGAM variants alter Mac-1 functions on neutrophils.Arthritis Rheum. 2013 Nov;65(11):2907-16. doi: 10.1002/art.38117. Arthritis Rheum. 2013. PMID: 23918739 Free PMC article.
-
The rs1143679 (R77H) lupus associated variant of ITGAM (CD11b) impairs complement receptor 3 mediated functions in human monocytes.Ann Rheum Dis. 2012 Dec;71(12):2028-34. doi: 10.1136/annrheumdis-2012-201390. Epub 2012 May 14. Ann Rheum Dis. 2012. PMID: 22586164 Free PMC article.
-
Phagocytosis is the main CR3-mediated function affected by the lupus-associated variant of CD11b in human myeloid cells.PLoS One. 2013;8(2):e57082. doi: 10.1371/journal.pone.0057082. Epub 2013 Feb 22. PLoS One. 2013. PMID: 23451151 Free PMC article.
-
The CD11b-integrin (ITGAM) and systemic lupus erythematosus.Lupus. 2013 Jun;22(7):657-63. doi: 10.1177/0961203313491851. Lupus. 2013. PMID: 23753600 Review.
-
Association between the functional ITGAM rs1143679 G/A polymorphism and systemic lupus erythematosus/lupus nephritis or rheumatoid arthritis: an update meta-analysis.Rheumatol Int. 2015 May;35(5):815-23. doi: 10.1007/s00296-014-3156-2. Epub 2014 Oct 16. Rheumatol Int. 2015. PMID: 25315704 Review.
Cited by
-
Systemic lupus erythematosus as a genetic disease.Clin Immunol. 2022 Mar;236:108953. doi: 10.1016/j.clim.2022.108953. Epub 2022 Feb 9. Clin Immunol. 2022. PMID: 35149194 Free PMC article.
-
Identification of Key Genes and Pathways in Myeloma side population cells by Bioinformatics Analysis.Int J Med Sci. 2020 Jul 25;17(14):2063-2076. doi: 10.7150/ijms.48244. eCollection 2020. Int J Med Sci. 2020. PMID: 32922167 Free PMC article.
-
Lupus Nephritis: Current Perspectives and Moving Forward.J Inflamm Res. 2022 Dec 2;15:6533-6552. doi: 10.2147/JIR.S363722. eCollection 2022. J Inflamm Res. 2022. PMID: 36483271 Free PMC article. Review.
-
Identification of crucial genes that induce coronary atherosclerosis through endothelial cell dysfunction in AMI-identifying hub genes by WGCNA.Am J Transl Res. 2022 Nov 15;14(11):8166-8174. eCollection 2022. Am J Transl Res. 2022. PMID: 36505315 Free PMC article.
-
De novo mutations implicate novel genes in systemic lupus erythematosus.Hum Mol Genet. 2018 Feb 1;27(3):421-429. doi: 10.1093/hmg/ddx407. Hum Mol Genet. 2018. PMID: 29177435 Free PMC article.
References
-
- Harley JB, Alarcón-Riquelme ME, Criswell L a, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD, Nath SK, Guthridge JM, Cobb BL, Mirel DB, Marion MC, Williams AH, Divers J, Wang W, Frank SG, Namjou B, Gabriel SB, Lee AT, Gregersen PK, Behrens TW, Taylor KE, Fernando M, Zidovetzki R, Gaffney PM, Edberg JC, Rioux JD, Ojwang JO. et al.Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. - DOI - PMC - PubMed
-
- Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, Mackay TFC, McCarroll SA, Visscher PM. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

