CRISPR/Cas9-mediated phage resistance is not impeded by the DNA modifications of phage T4
- PMID: 24886988
- PMCID: PMC4041780
- DOI: 10.1371/journal.pone.0098811
CRISPR/Cas9-mediated phage resistance is not impeded by the DNA modifications of phage T4
Abstract
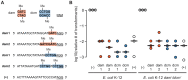
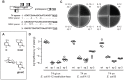
Bacteria rely on two known DNA-level defenses against their bacteriophage predators: restriction-modification and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-CRISPR-associated (Cas) systems. Certain phages have evolved countermeasures that are known to block endonucleases. For example, phage T4 not only adds hydroxymethyl groups to all of its cytosines, but also glucosylates them, a strategy that defeats almost all restriction enzymes. We sought to determine whether these DNA modifications can similarly impede CRISPR-based defenses. In a bioinformatics search, we found naturally occurring CRISPR spacers that potentially target phages known to modify their DNA. Experimentally, we show that the Cas9 nuclease from the Type II CRISPR system of Streptococcus pyogenes can overcome a variety of DNA modifications in Escherichia coli. The levels of Cas9-mediated phage resistance to bacteriophage T4 and the mutant phage T4 gt, which contains hydroxymethylated but not glucosylated cytosines, were comparable to phages with unmodified cytosines, T7 and the T4-like phage RB49. Our results demonstrate that Cas9 is not impeded by N6-methyladenine, 5-methylcytosine, 5-hydroxymethylated cytosine, or glucosylated 5-hydroxymethylated cytosine.
Conflict of interest statement
Figures


Similar articles
-
Covalent Modification of Bacteriophage T4 DNA Inhibits CRISPR-Cas9.mBio. 2015 Jun 16;6(3):e00648. doi: 10.1128/mBio.00648-15. mBio. 2015. PMID: 26081634 Free PMC article.
-
Covalent Modifications of the Bacteriophage Genome Confer a Degree of Resistance to Bacterial CRISPR Systems.J Virol. 2020 Nov 9;94(23):e01630-20. doi: 10.1128/JVI.01630-20. Print 2020 Nov 9. J Virol. 2020. PMID: 32938767 Free PMC article.
-
Bacteriophage DNA glucosylation impairs target DNA binding by type I and II but not by type V CRISPR-Cas effector complexes.Nucleic Acids Res. 2018 Jan 25;46(2):873-885. doi: 10.1093/nar/gkx1264. Nucleic Acids Res. 2018. PMID: 29253268 Free PMC article.
-
Protein Inhibitors of CRISPR-Cas9.ACS Chem Biol. 2018 Feb 16;13(2):417-423. doi: 10.1021/acschembio.7b00831. Epub 2018 Jan 17. ACS Chem Biol. 2018. PMID: 29251498 Free PMC article. Review.
-
Molecular mechanisms of CRISPR-mediated microbial immunity.Cell Mol Life Sci. 2014 Feb;71(3):449-65. doi: 10.1007/s00018-013-1438-6. Cell Mol Life Sci. 2014. PMID: 23959171 Free PMC article. Review.
Cited by
-
Hypermodified DNA in Viruses of E. coli and Salmonella.EcoSal Plus. 2021 Dec 15;9(2):eESP00282019. doi: 10.1128/ecosalplus.ESP-0028-2019. Epub 2021 Sep 28. EcoSal Plus. 2021. PMID: 34910575 Free PMC article. Review.
-
Optimization of T4 phage engineering via CRISPR/Cas9.Sci Rep. 2020 Oct 26;10(1):18229. doi: 10.1038/s41598-020-75426-6. Sci Rep. 2020. PMID: 33106580 Free PMC article.
-
Emergent rules for codon choice elucidated by editing rare arginine codons in Escherichia coli.Proc Natl Acad Sci U S A. 2016 Sep 20;113(38):E5588-97. doi: 10.1073/pnas.1605856113. Epub 2016 Sep 6. Proc Natl Acad Sci U S A. 2016. PMID: 27601680 Free PMC article.
-
Covalent Modification of Bacteriophage T4 DNA Inhibits CRISPR-Cas9.mBio. 2015 Jun 16;6(3):e00648. doi: 10.1128/mBio.00648-15. mBio. 2015. PMID: 26081634 Free PMC article.
-
Complete Genome Sequences of T4-Like Bacteriophages RB3, RB5, RB6, RB7, RB9, RB10, RB27, RB33, RB55, RB59, and RB68.Genome Announc. 2015 Jan 2;3(1):e01122-14. doi: 10.1128/genomeA.01122-14. Genome Announc. 2015. PMID: 25555735 Free PMC article.
References
-
- Lehman IR, Pratt EA (1960) On the structure of the glucosylated hydroxymethylcytosine nucleotides of coliphages T2, T4, and T6. J Biol Chem 235: 3254–3259. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

