Genome size variation and evolution in allotetraploid Arabidopsis kamchatica and its parents, Arabidopsis lyrata and Arabidopsis halleri
- PMID: 24887004
- PMCID: PMC4076644
- DOI: 10.1093/aobpla/plu025
Genome size variation and evolution in allotetraploid Arabidopsis kamchatica and its parents, Arabidopsis lyrata and Arabidopsis halleri
Abstract
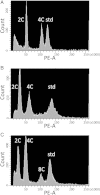
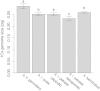
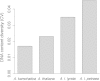
Polyploidization and subsequent changes in genome size are fundamental processes in evolution and diversification. Little is currently known about the extent of genome size variation within taxa and the evolutionary forces acting on this variation. Arabidopsis kamchatica has been reported to contain both diploid and tetraploid individuals. The aim of this study was to determine the genome size of A. kamchatica, whether there is variation in ploidy and/or genome size in A. kamchatica and to study how genome size has evolved. We used propidium iodide flow cytometry to measure 2C DNA content of 73 plants from 25 geographically diverse populations of the putative allotetraploid A. kamchatica and its parents, Arabidopsis lyrata and Arabidopsis halleri. All A. kamchatica plants appear to be tetraploids. The mean 2C DNA content of A. kamchatica was 1.034 pg (1011 Mbp), which is slightly smaller than the sum of its diploid parents (A. lyrata: 0.502 pg; A. halleri: 0.571 pg). Arabidopsis kamchatica appears to have lost ∼37.594 Mbp (3.6 %) of DNA from its 2C genome. Tetraploid A. lyrata from Germany and Austria appears to have lost ∼70.366 Mbp (7.2 %) of DNA from the 2C genome, possibly due to hybridization with A. arenosa, which has a smaller genome than A. lyrata. We did find genome size differences among A. kamchatica populations, which varied up to 7 %. Arabidopsis kamchatica ssp. kawasakiana from Japan appears to have a slightly larger genome than A. kamchatica ssp. kamchatica from North America, perhaps due to multiple allopolyploid origins or hybridization with A. halleri. However, the among-population coefficient of variation in 2C DNA content is lower in A. kamchatica than in other Arabidopsis taxa. Due to its close relationship to A. thaliana, A. kamchatica has the potential to be very useful in the study of polyploidy and genome evolution.
Keywords: 2C DNA content; Allotetraploid; Arabidopsis halleri ssp. gemmifera; Arabidopsis kamchatica; Arabidopsis lyrata; C-value; flow cytometry; genome size; genome size variation.
Published by Oxford University Press on behalf of the Annals of Botany Company.
Figures

Similar articles
-
The allopolyploid Arabidopsis kamchatica originated from multiple individuals of Arabidopsis lyrata and Arabidopsis halleri.Mol Ecol. 2009 Oct;18(19):4024-48. doi: 10.1111/j.1365-294X.2009.04329.x. Epub 2009 Sep 15. Mol Ecol. 2009. PMID: 19754506
-
Conserved but Attenuated Parental Gene Expression in Allopolyploids: Constitutive Zinc Hyperaccumulation in the Allotetraploid Arabidopsis kamchatica.Mol Biol Evol. 2016 Nov;33(11):2781-2800. doi: 10.1093/molbev/msw141. Epub 2016 Jul 12. Mol Biol Evol. 2016. PMID: 27413047 Free PMC article.
-
The evolutionary history of the Arabidopsis lyrata complex: a hybrid in the amphi-Beringian area closes a large distribution gap and builds up a genetic barrier.BMC Evol Biol. 2010 Apr 8;10:98. doi: 10.1186/1471-2148-10-98. BMC Evol Biol. 2010. PMID: 20377907 Free PMC article.
-
Polyploid Arabidopsis species originated around recent glaciation maxima.Curr Opin Plant Biol. 2018 Apr;42:8-15. doi: 10.1016/j.pbi.2018.01.005. Epub 2018 Feb 12. Curr Opin Plant Biol. 2018. PMID: 29448159 Review.
-
Poorly known relatives of Arabidopsis thaliana.Trends Plant Sci. 2006 Sep;11(9):449-59. doi: 10.1016/j.tplants.2006.07.005. Epub 2006 Aug 8. Trends Plant Sci. 2006. PMID: 16893672 Review.
Cited by
-
The impacts of polyploidy, geographic and ecological isolations on the diversification of Panax (Araliaceae).BMC Plant Biol. 2015 Dec 21;15:297. doi: 10.1186/s12870-015-0669-0. BMC Plant Biol. 2015. PMID: 26690782 Free PMC article.
-
Taming the wild: resolving the gene pools of non-model Arabidopsis lineages.BMC Evol Biol. 2014 Oct 27;14:224. doi: 10.1186/s12862-014-0224-x. BMC Evol Biol. 2014. PMID: 25344686 Free PMC article.
-
Unraveling the maternal and paternal origins of allotetraploid Vigna reflexo-pilosa.Sci Rep. 2023 Dec 22;13(1):22951. doi: 10.1038/s41598-023-49908-2. Sci Rep. 2023. PMID: 38135720 Free PMC article.
References
-
- Adams KL, Wendel JF. Polyploidy and genome evolution in plants. Current Opinion in Plant Biology. 2005;8:135–141. - PubMed
-
- Angulo MB, Dematteis M. Nuclear DNA content in some species of Lessingianthus (Vernonieae, Asteraceae) by flow cytometry. Journal of Plant Research. 2013;126:461–468. - PubMed
-
- Bates D. Fitting linear mixed models in R. R News. 2005;5:27–30.
-
- Bennett MD. Nuclear DNA content and minimum generation time in herbaceous plants. Proceedings of the Royal Society B Biological Sciences. 1972;181:109–135. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

