A Gondwanan imprint on global diversity and domestication of wine and cider yeast Saccharomyces uvarum
- PMID: 24887054
- PMCID: PMC5081218
- DOI: 10.1038/ncomms5044
A Gondwanan imprint on global diversity and domestication of wine and cider yeast Saccharomyces uvarum
Abstract
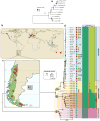
In addition to Saccharomyces cerevisiae, the cryotolerant yeast species S. uvarum is also used for wine and cider fermentation but nothing is known about its natural history. Here we use a population genomics approach to investigate its global phylogeography and domestication fingerprints using a collection of isolates obtained from fermented beverages and from natural environments on five continents. South American isolates contain more genetic diversity than that found in the Northern Hemisphere. Moreover, coalescence analyses suggest that a Patagonian sub-population gave rise to the Holarctic population through a recent bottleneck. Holarctic strains display multiple introgressions from other Saccharomyces species, those from S. eubayanus being prevalent in European strains associated with human-driven fermentations. These introgressions are absent in the large majority of wild strains and gene ontology analyses indicate that several gene categories relevant for wine fermentation are overrepresented. Such findings constitute a first indication of domestication in S. uvarum.
Figures
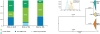

References
-
- Cavalieri D, McGovern PE, Hartl DL, Mortimer R, Polsineli M. Evidence for S. cerevisiae fermentation in ancient wine. J Mol Evol. 2003;57(Suppl 1):S226–S232. - PubMed
-
- Hittinger CT. Saccharomyces diversity and evolution: a budding model genus. Trends Genet. 2013;29:309–317. - PubMed
-
- Wang QM, Liu WQ, Liti G, Wang SA, Bai FY. Surprisingly diverged populations of Saccharomyces cerevisiae in natural environments remote from human activity. Mol Ecol. 2012;21:5404–5417. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

