Changes in abundance of oral microbiota associated with oral cancer
- PMID: 24887397
- PMCID: PMC4041887
- DOI: 10.1371/journal.pone.0098741
Changes in abundance of oral microbiota associated with oral cancer
Erratum in
- PLoS One. 2014;9(8):e106297. Muy-Teck The [removed]
Abstract
Individual bacteria and shifts in the composition of the microbiome have been associated with human diseases including cancer. To investigate changes in the microbiome associated with oral cancers, we profiled cancers and anatomically matched contralateral normal tissue from the same patient by sequencing 16S rDNA hypervariable region amplicons. In cancer samples from both a discovery and a subsequent confirmation cohort, abundance of Firmicutes (especially Streptococcus) and Actinobacteria (especially Rothia) was significantly decreased relative to contralateral normal samples from the same patient. Significant decreases in abundance of these phyla were observed for pre-cancers, but not when comparing samples from contralateral sites (tongue and floor of mouth) from healthy individuals. Weighted UniFrac principal coordinates analysis based on 12 taxa separated most cancers from other samples with greatest separation of node positive cases. These studies begin to develop a framework for exploiting the oral microbiome for monitoring oral cancer development, progression and recurrence.
Conflict of interest statement
Figures


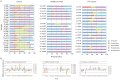
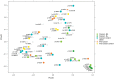
References
-
- Parkin DM, Pisani P, Ferlay J (1999) Global cancer statistics. CA Cancer J Clin 49: 33–64, 31. - PubMed
-
- Shiboski CH, Schmidt BL, Jordan RC (2005) Tongue and tonsil carcinoma: increasing trends in the U.S. population ages 20–44 years. Cancer 103: 1843–1849. - PubMed
-
- Schmidt BL, Dierks EJ, Homer L, Potter B (2004) Tobacco smoking history and presentation of oral squamous cell carcinoma. J Oral Maxillofac Surg 62: 1055–1058. - PubMed
-
- Gillison ML (2004) Human papillomavirus-associated head and neck cancer is a distinct epidemiologic, clinical, and molecular entity. Semin Oncol 31: 744–754. - PubMed
-
- Correa P, Haenszel W, Cuello C, Tannenbaum S, Archer M (1975) A model for gastric cancer epidemiology. Lancet 2: 58–60. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

