Enzyme structure captures four cysteines aligned for disulfide relay
- PMID: 24888638
- PMCID: PMC4116658
- DOI: 10.1002/pro.2496
Enzyme structure captures four cysteines aligned for disulfide relay
Abstract
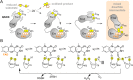
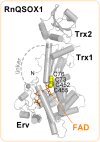
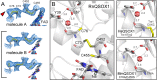
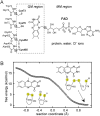
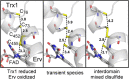
Thioredoxin superfamily proteins introduce disulfide bonds into substrates, catalyze the removal of disulfides, and operate in electron relays. These functions rely on one or more dithiol/disulfide exchange reactions. The flavoenzyme quiescin sulfhydryl oxidase (QSOX), a catalyst of disulfide bond formation with an interdomain electron transfer step in its catalytic cycle, provides a unique opportunity for exploring the structural environment of enzymatic dithiol/disulfide exchange. Wild-type Rattus norvegicus QSOX1 (RnQSOX1) was crystallized in a conformation that juxtaposes the two redox-active di-cysteine motifs in the enzyme, presenting the entire electron-transfer pathway and proton-transfer participants in their native configurations. As such a state cannot generally be enriched and stabilized for analysis, RnQSOX1 gives unprecedented insight into the functional group environments of the four cysteines involved in dithiol/disulfide exchange and provides the framework for analysis of the energetics of electron transfer in the presence of the bound flavin adenine dinucleotide cofactor. Hybrid quantum mechanics/molecular mechanics (QM/MM) free energy simulations based on the X-ray crystal structure suggest that formation of the interdomain disulfide intermediate is highly favorable and secures the flexible enzyme in a state from which further electron transfer via the flavin can occur.
Keywords: X-ray crystallography; cis-proline; enzyme mechanism; flavin adenine dinucleotide; quantum mechanics/molecular mechanics; thioredoxin fold.
© 2014 The Protein Society.
Figures

References
-
- Arnér ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–6109. - PubMed
-
- Heras B, Shouldice SR, Totsika M, Scanlon MJ, Schembri MA, Martin JL. DSB proteins and bacterial pathogenicity. Nat Rev Microbiol. 2009;7:215–225. - PubMed
-
- Bulleid N, Ellgaard L. Multiple ways to make disulfides. Trends Biochem Sci. 2011;36:485–492. - PubMed
-
- Martin JL. Thioredoxin—a fold for all reasons. Structure. 1995;3:245–250. - PubMed
-
- Chivers PT, Prehoda KE, Raines RT. The CXXC motif: a rheostat in the active site. Biochemistry. 1997;36:4061–4066. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

