Monitoring the dynamics of clonal tumour evolution in vivo using secreted luciferases
- PMID: 24889111
- PMCID: PMC4059931
- DOI: 10.1038/ncomms4981
Monitoring the dynamics of clonal tumour evolution in vivo using secreted luciferases
Abstract
Tumours are heterogeneous cell populations that undergo clonal evolution during tumour progression, metastasis and response to therapy. Short hairpin RNAs (shRNAs) generate stable loss-of-function phenotypes and are versatile experimental tools to explore the contribution of individual genetic alterations to clonal evolution. In these experiments tumour cells carrying shRNAs are commonly tracked with fluorescent reporters. While this works well for cell culture studies and leukaemia mouse models, fluorescent reporters are poorly suited for animals with solid tumours--the most common tumour types in cancer patients. Here we develop a toolkit that uses secreted luciferases to track the fate of two different shRNA-expressing tumour cell clones competitively, both in vitro and in vivo. We demonstrate that secreted luciferase activities can be measured robustly in the blood stream of tumour-bearing mice to accurately quantify, in a minimally invasive manner, the dynamic evolution of two genetically distinct tumour subclones in preclinical mouse models of tumour development, metastasis and therapy.
Figures



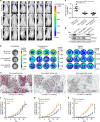
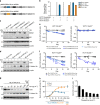

References
-
- Cairns J. Mutation selection and the natural history of cancer. Nature 255, 197–200 (1975). - PubMed
-
- Nowell P. C. The clonal evolution of tumor cell populations. Science 194, 23–28 (1976). - PubMed
-
- Burrell R. A., McGranahan N., Bartek J. & Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 501, 338–345 (2013). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

