A new sensitive PCR assay for one-step detection of 12 IDH1/2 mutations in glioma
- PMID: 24889502
- PMCID: PMC4229941
- DOI: 10.1186/2051-5960-2-58
A new sensitive PCR assay for one-step detection of 12 IDH1/2 mutations in glioma
Abstract
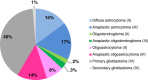
Introduction: Mutations in isocitrate dehydrogenase genes IDH1 or IDH2 are frequent in glioma, and IDH mutation status is a strong diagnostic and prognostic marker. Current IDH mutation screening is performed with an immunohistochemistry (IHC) assay specific for IDH1 R132H, the most common mutation. Sequencing is recommended as a second-step test for IHC-negative or -equivocal cases. We developed and validated a new real-time quantitative polymerase chain reaction (PCR) assay for single-step detection of IDH1 R132H and 11 rare IDH1/2 mutations in formalin-fixed paraffin-embedded (FFPE) glioma samples. Performance of the IDH1/2 PCR assay was compared to IHC and Sanger sequencing.
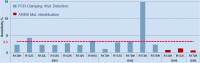
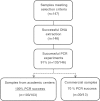
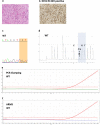
Results: The IDH1/2 PCR assay combines PCR clamping for detection of 7 IDH1 and 5 IDH2 mutations, and Amplification Refractory Mutation System technology for specific identification of the 3 most common mutations (IDH1 R132H, IDH1 R132C, IDH2 R172K). Analytical sensitivity of the PCR assay for mutation detection was <5% for 11/12 mutations (mean: 3.3%), and sensitivity for mutation identification was very high (0.8% for IDH1 R132H; 1.2% for IDH1 R132C; 0.6% for IDH2 R172K). Assay performance was further validated on 171 clinical glioma FFPE samples; of these, 147 samples met the selection criteria and 146 DNA samples were successfully extracted. IDH1/2 status was successfully obtained in 91% of cases. All but one positive IDH1 R132H-IHC cases were concordantly detected by PCR and 3 were not detected by sequencing. Among the IHC-negative cases (n = 72), PCR detected 12 additional rare mutations (10 IDH1, 2 IDH2). All mutations detected by sequencing (n = 67) were concordantly detected by PCR and 5/66 sequencing-negative cases were PCR-positive (overall concordance: 96%). Analysis of synthetic samples representative of the 11 rare IDH1/2 mutations detected by the assay produced 100% correct results.
Conclusions: The new IDH1/2 PCR assay has a high technical success rate and is more sensitive than Sanger sequencing. Positive concordance was 98% with IHC for IDH1 R132H detection and 100% with sequencing. The PCR assay can reliably be performed on FFPE samples and has a faster turnaround time than current IDH mutation detection algorithms. The assay should facilitate implementation of a comprehensive IDH1/2 testing protocol in routine clinical practice.
Figures

References
-
- Hartmann C, Meyer J, Balss J, Capper D, Mueller W, Christians A, Felsberg J, Wolter M, Mawrin C, Wick W, Weller M, Herold-Mende C, Unterberg A, Jeuken JW, Wesseling P, Reifenberger G, von Deimling A. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118(4):469–474. doi: 10.1007/s00401-009-0561-9. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

