Imaging genomic mapping of an invasive MRI phenotype predicts patient outcome and metabolic dysfunction: a TCGA glioma phenotype research group project
- PMID: 24889866
- PMCID: PMC4057583
- DOI: 10.1186/1755-8794-7-30
Imaging genomic mapping of an invasive MRI phenotype predicts patient outcome and metabolic dysfunction: a TCGA glioma phenotype research group project
Abstract
Background: Invasion of tumor cells into adjacent brain parenchyma is a major cause of treatment failure in glioblastoma. Furthermore, invasive tumors are shown to have a different genomic composition and metabolic abnormalities that allow for a more aggressive GBM phenotype and resistance to therapy. We thus seek to identify those genomic abnormalities associated with a highly aggressive and invasive GBM imaging-phenotype.
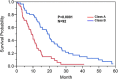
Methods: We retrospectively identified 104 treatment-naïve glioblastoma patients from The Cancer Genome Atlas (TCGA) whom had gene expression profiles and corresponding MR imaging available in The Cancer Imaging Archive (TCIA). The standardized VASARI feature-set criteria were used for the qualitative visual assessments of invasion. Patients were assigned to classes based on the presence (Class A) or absence (Class B) of statistically significant invasion parameters to create an invasive imaging signature; imaging genomic analysis was subsequently performed using GenePattern Comparative Marker Selection module (Broad Institute).
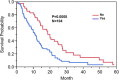
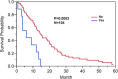
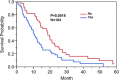
Results: Our results show that patients with a combination of deep white matter tracts and ependymal invasion (Class A) on imaging had a significant decrease in overall survival as compared to patients with absence of such invasive imaging features (Class B) (8.7 versus 18.6 months, p < 0.001). Mitochondrial dysfunction was the top canonical pathway associated with Class A gene expression signature. The MYC oncogene was predicted to be the top activation regulator in Class A.
Conclusion: We demonstrate that MRI biomarker signatures can identify distinct GBM phenotypes associated with highly significant survival differences and specific molecular pathways. This study identifies mitochondrial dysfunction as the top canonical pathway in a very aggressive GBM phenotype. Thus, imaging-genomic analyses may prove invaluable in detecting novel targetable genomic pathways.
Figures






References
-
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;7:987–996. doi: 10.1056/NEJMoa043330. - DOI - PubMed
-
- Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, Phillimore B, Begum S, McDonald NQ, Butler A, Jones D, Raine K, Latimer C, Santos CR, Nohadani M, Eklund AC, Spencer-Dene B, Clark G, Pickering L, Stamp G, Gore M, Szallasi Z, Downward J, Futreal PA, Swanton C. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;7:883–892. doi: 10.1056/NEJMoa1113205. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

