Origin of spliceosomal introns and alternative splicing
- PMID: 24890509
- PMCID: PMC4031966
- DOI: 10.1101/cshperspect.a016071
Origin of spliceosomal introns and alternative splicing
Abstract
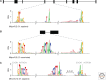
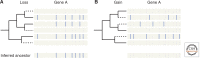
In this work we review the current knowledge on the prehistory, origins, and evolution of spliceosomal introns. First, we briefly outline the major features of the different types of introns, with particular emphasis on the nonspliceosomal self-splicing group II introns, which are widely thought to be the ancestors of spliceosomal introns. Next, we discuss the main scenarios proposed for the origin and proliferation of spliceosomal introns, an event intimately linked to eukaryogenesis. We then summarize the evidence that suggests that the last eukaryotic common ancestor (LECA) had remarkably high intron densities and many associated characteristics resembling modern intron-rich genomes. From this intron-rich LECA, the different eukaryotic lineages have taken very distinct evolutionary paths leading to profoundly diverged modern genome structures. Finally, we discuss the origins of alternative splicing and the qualitative differences in alternative splicing forms and functions across lineages.
Copyright © 2014 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures


References
-
- Abramovitz DL, Friedman RA, Pyle AM 1996. Catalytic role of 2′-hydroxyl groups within a group II intron active site. Science 271: 1410–1413 - PubMed
-
- Adams KL, Palmer JD 2003. Evolution of mitochondrial gene content: Gene loss and transfer to the nucleus. Mol Phylogenet Evol 29: 380–395 - PubMed
-
- Amrani N, Ganesan R, Kervestin S, Mangus DA, Ghosh S, Jacobson A 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432: 112–118 - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
