CSF-1 receptor signaling in myeloid cells
- PMID: 24890514
- PMCID: PMC4031967
- DOI: 10.1101/cshperspect.a021857
CSF-1 receptor signaling in myeloid cells
Abstract
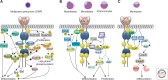
The CSF-1 receptor (CSF-1R) is activated by the homodimeric growth factors colony-stimulating factor-1 (CSF-1) and interleukin-34 (IL-34). It plays important roles in development and in innate immunity by regulating the development of most tissue macrophages and osteoclasts, of Langerhans cells of the skin, of Paneth cells of the small intestine, and of brain microglia. It also regulates the differentiation of neural progenitor cells and controls functions of oocytes and trophoblastic cells in the female reproductive tract. Owing to this broad tissue expression pattern, it plays a central role in neoplastic, inflammatory, and neurological diseases. In this review we summarize the evolution, structure, and regulation of expression of the CSF-1R gene. We discuss the structures of CSF-1, IL-34, and the CSF-1R and the mechanism of ligand binding to and activation of the receptor. We further describe the pathways regulating macrophage survival, proliferation, differentiation, and chemotaxis downstream from the CSF-1R.
Copyright © 2014 Cold Spring Harbor Laboratory Press; all rights reserved.
Figures




References
-
- Baccarini M, Li W, Dello Sbarba P, Stanley ER 1991. Increased phosphorylation of the colony stimulating factor-1 receptor following transmembrane signaling. Receptor 1: 243–259 - PubMed
-
- Baran CP, Tridandapani S, Helgason CD, Humphries RK, Krystal G, Marsh CB 2003. The inositol 5′-phosphatase SHIP-1 and the Src kinase Lyn negatively regulate macrophage colony-stimulating factor-induced Akt activity. J Biol Chem 278: 38628–38636 - PubMed
-
- Bartelmez SH, Stanley ER 1985. Synergism between hemopoietic growth factors (HGFs) detected by their effects on cells bearing receptors for a lineage specific HGF: Assay of hemopoietin-1. J Cell Physiol 122: 370–378 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
