Characterization of conventional and atypical receptors for the chemokine CCL2 on mouse leukocytes
- PMID: 24890717
- PMCID: PMC4065784
- DOI: 10.4049/jimmunol.1303236
Characterization of conventional and atypical receptors for the chemokine CCL2 on mouse leukocytes
Abstract
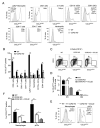
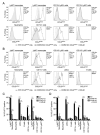
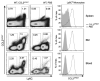
Chemokine-directed leukocyte migration is crucial for effective immune and inflammatory responses. Conventional chemokine receptors (cCKRs) directly control cell movement; atypical chemokine receptors (ACKRs) regulate coexpressed cCKRs; and both cCKRs and ACKRs internalize chemokines to limit their abundance in vivo, a process referred to as scavenging. A leukocyte's migratory and chemokine-scavenging potential is determined by which cCKRs and ACKRs it expresses, and by the ligand specificity, signaling properties, and chemokine internalization capacity of these receptors. Most chemokines can bind at least one cCKR and one ACKR. CCL2 can bind to CCR2 (a cCKR) and two ACKRs (ACKR1 and ACKR2). In this study, by using fluorescent CCL2 uptake to label cells bearing functional CCL2 receptors, we have defined the expression profile, scavenging activity, and ligand specificity of CCL2 receptors on mouse leukocytes. We show that qualitative and quantitative differences in the expression of CCR2 and ACKR2 endow individual leukocyte subsets with distinctive CCL2 receptor profiles and CCL2-scavenging capacities. We reveal that some cells, including plasmacytoid dendritic cells, can express both CCR2 and ACKR2; that Ly6C(high) monocytes have particularly strong CCL2-scavenging potential in vitro and in vivo; and that CCR2 is a much more effective CCL2 scavenger than ACKR2. We confirm the unique, overlapping, ligand specificities of CCR2 and ACKR2 and, unexpectedly, find that cell context influences the interaction of CCL7 and CCL12 with CCR2. Fluorescent chemokine uptake assays were instrumental in providing these novel insights into CCL2 receptor biology, and the sensitivity, specificity, and versatility of these assays are discussed.
Copyright © 2014 by The American Association of Immunologists, Inc.
Figures

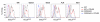
Similar articles
-
Atypical chemokine receptor ACKR2 mediates chemokine scavenging by primary human trophoblasts and can regulate fetal growth, placental structure, and neonatal mortality in mice.J Immunol. 2014 Nov 15;193(10):5218-28. doi: 10.4049/jimmunol.1401096. Epub 2014 Oct 8. J Immunol. 2014. PMID: 25297873
-
The dual-function chemokine receptor CCR2 drives migration and chemokine scavenging through distinct mechanisms.Sci Signal. 2023 Jan 31;16(770):eabo4314. doi: 10.1126/scisignal.abo4314. Epub 2023 Jan 31. Sci Signal. 2023. PMID: 36719944 Free PMC article.
-
The CCL2/CCL7/CCL12/CCR2 pathway is substantially and persistently upregulated in mice after traumatic brain injury, and CCL2 modulates the complement system in microglia.Mol Cell Probes. 2020 Dec;54:101671. doi: 10.1016/j.mcp.2020.101671. Epub 2020 Nov 4. Mol Cell Probes. 2020. PMID: 33160071
-
A CCL2-based fusokine as a novel biopharmaceutical for the treatment of CCR2-driven autoimmune diseases.Crit Rev Immunol. 2010;30(5):449-61. doi: 10.1615/critrevimmunol.v30.i5.40. Crit Rev Immunol. 2010. PMID: 21083526 Review.
-
Beyond Cell Motility: The Expanding Roles of Chemokines and Their Receptors in Malignancy.Front Immunol. 2020 Jun 4;11:952. doi: 10.3389/fimmu.2020.00952. eCollection 2020. Front Immunol. 2020. PMID: 32582148 Free PMC article. Review.
Cited by
-
A Fluorescent Activatable AND-Gate Chemokine CCL2 Enables In Vivo Detection of Metastasis-Associated Macrophages.Angew Chem Int Ed Engl. 2019 Nov 18;58(47):16894-16898. doi: 10.1002/anie.201910955. Epub 2019 Oct 11. Angew Chem Int Ed Engl. 2019. PMID: 31535788 Free PMC article.
-
Carcinoembryonic Antigen Cell Adhesion Molecule 1 long isoform modulates malignancy of poorly differentiated colon cancer cells.Gut. 2016 May;65(5):821-9. doi: 10.1136/gutjnl-2014-308781. Epub 2015 Feb 9. Gut. 2016. PMID: 25666195 Free PMC article.
-
Hypertrophic heart failure promotes gut dysbiosis and gut leakage in interleukin 10-deficient mice.Am J Physiol Heart Circ Physiol. 2025 Mar 1;328(3):H447-H459. doi: 10.1152/ajpheart.00323.2024. Epub 2025 Jan 24. Am J Physiol Heart Circ Physiol. 2025. PMID: 39854049 Free PMC article.
-
Inhibition of CCL2 by bindarit alleviates diabetes-associated periodontitis by suppressing inflammatory monocyte infiltration and altering macrophage properties.Cell Mol Immunol. 2021 Sep;18(9):2224-2235. doi: 10.1038/s41423-020-0500-1. Epub 2020 Jul 16. Cell Mol Immunol. 2021. PMID: 32678310 Free PMC article.
-
Nonmotile Single-Cell Migration as a Random Walk in Nonuniformity: The "Extreme Dumping Limit" for Cell-to-Cell Communications.J Healthc Eng. 2018 Nov 25;2018:9680713. doi: 10.1155/2018/9680713. eCollection 2018. J Healthc Eng. 2018. PMID: 30595832 Free PMC article.
References
-
- Bachelerie F, Ben-Baruch A, Burkhardt AM, Combadière C, Farber JM, Graham GJ, Horuk R, Sparre-Ulrich AH, Locati M, Luster AD, Mantovani A, Matsushima K, Murphy PM, Nibbs R, Nomiyama H, Power CA, Proudfoot AEI, Rosenkilde MM, Rot A, Sozzani S, Thelen M, Yoshie O, Zlotnik A. International Union of Pharmacology. LXXXIX. Update on the Extended Family of Chemokine Receptors and Introducing a New Nomenclature for Atypical Chemokine Receptors. Pharmacol Rev. 2014;66:1–79. - PMC - PubMed
-
- Nibbs RJB, Graham GJ. Immune regulation by atypical chemokine receptors. Nat Rev Immunol. 2013;13:815–829. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

