Effects of new mutations on fitness: insights from models and data
- PMID: 24891070
- PMCID: PMC4282485
- DOI: 10.1111/nyas.12460
Effects of new mutations on fitness: insights from models and data
Abstract
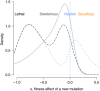
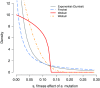
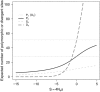
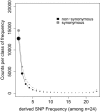
The rates and properties of new mutations affecting fitness have implications for a number of outstanding questions in evolutionary biology. Obtaining estimates of mutation rates and effects has historically been challenging, and little theory has been available for predicting the distribution of fitness effects (DFE); however, there have been recent advances on both fronts. Extreme-value theory predicts the DFE of beneficial mutations in well-adapted populations, while phenotypic fitness landscape models make predictions for the DFE of all mutations as a function of the initial level of adaptation and the strength of stabilizing selection on traits underlying fitness. Direct experimental evidence confirms predictions on the DFE of beneficial mutations and favors distributions that are roughly exponential but bounded on the right. A growing number of studies infer the DFE using genomic patterns of polymorphism and divergence, recovering a wide range of DFE. Future work should be aimed at identifying factors driving the observed variation in the DFE. We emphasize the need for further theory explicitly incorporating the effects of partial pleiotropy and heterogeneity in the environment on the expected DFE.
Keywords: distribution of fitness effects; experimental evolution; mutation; mutational landscape models; population genomics.
© 2014 The Authors. Annals of the New York Academy of Sciences published by Wiley Periodicals Inc. on behalf of The New York Academy of Sciences.
Figures
References
-
- Chevin L-M, Martin G. Lenormand T. Fisher's model and the genomics of adaptation: restricted pleiotropy, heterogeneous mutation, and parallel evolution. Evolution. 2010;64:3213–3231. - PubMed
-
- Hoffmann AA. Sgrò CM. Climate change and evolutionary adaptation. Nature. 2011;470:479–485. - PubMed
-
- Otto SP. Lenormand T. Resolving the paradox of sex and recombination. Nat. Rev. Genet. 2002;3:252–261. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

