Continuing challenges in influenza
- PMID: 24891213
- PMCID: PMC4159436
- DOI: 10.1111/nyas.12462
Continuing challenges in influenza
Abstract
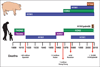
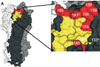
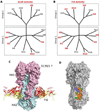
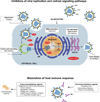
Influenza is an acute respiratory disease in mammals and domestic poultry that emerges from zoonotic reservoirs in aquatic birds and bats. Although influenza viruses are among the most intensively studied pathogens, existing control options require further improvement. Influenza vaccines must be regularly updated because of continuous antigenic drift and sporadic antigenic shifts in the viral surface glycoproteins. Currently, influenza therapeutics are limited to neuraminidase inhibitors; novel drugs and vaccine approaches are therefore urgently needed. Advances in vaccinology and structural analysis have revealed common antigenic epitopes on hemagglutinins across all influenza viruses and suggest that a universal influenza vaccine is possible. In addition, various immunomodulatory agents and signaling pathway inhibitors are undergoing preclinical development. Continuing challenges in influenza include the emergence of pandemic H1N1 influenza in 2009, human infections with avian H7N9 influenza in 2013, and sporadic human cases of highly pathogenic avian H5N1 influenza. Here, we review the challenges facing influenza scientists and veterinary and human public health officials; we also discuss the exciting possibility of achieving the ultimate goal of controlling influenza's ability to change its antigenicity.
Keywords: H1N1; H5N1; H7N9; antigenic changes; antiviral drugs; influenza virus; pandemic; vaccines; zoonosis.
© 2014 New York Academy of Sciences.
Figures

References
-
- Shaw ML, Palese P. Orthomyxoviradae: the viruses and their replication. In: Knipe D, Howley P, editors. Fields’ Virology, Sixth Edition. Philadelphia, PA: Lippin Williams & Wilkins; 2011. pp. 1647–1689.
-
- Jernigan DB, Cox NJ. Human influenza: One Health, One World. In: Webster RG, Monto AS, Braciale TJ, Lamb RA, editors. Textbook of Influenza, Second Edition. West Sussex: John Wiley and Sons; 2013. pp. 3–19.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

