Oxidation of cellular amino acid pools leads to cytotoxic mistranslation of the genetic code
- PMID: 24891238
- PMCID: PMC4066437
- DOI: 10.7554/eLife.02501
Oxidation of cellular amino acid pools leads to cytotoxic mistranslation of the genetic code
Abstract
Aminoacyl-tRNA synthetases use a variety of mechanisms to ensure fidelity of the genetic code and ultimately select the correct amino acids to be used in protein synthesis. The physiological necessity of these quality control mechanisms in different environments remains unclear, as the cost vs benefit of accurate protein synthesis is difficult to predict. We show that in Escherichia coli, a non-coded amino acid produced through oxidative damage is a significant threat to the accuracy of protein synthesis and must be cleared by phenylalanine-tRNA synthetase in order to prevent cellular toxicity caused by mis-synthesized proteins. These findings demonstrate how stress can lead to the accumulation of non-canonical amino acids that must be excluded from the proteome in order to maintain cellular viability.
Keywords: E. coli; S. cerevisiae; biochemistry; infectious disease; microbiology; protein synthesis; stress; translation.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures
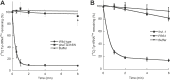
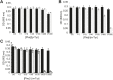


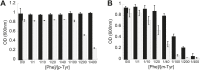



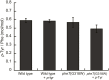

References
-
- Bertin C, Weston LA, Huang T, Jander G, Owens T, Meinwald J, Schroeder FC. 2007. Grass roots chemistry: meta-tyrosine, an herbicidal nonprotein amino acid. Proceedings of the National Academy of Sciences of the United States of America 104:16964–16969. doi: 10.1073/pnas.0707198104 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

