Localization of the outer membrane protein OmpA2 in Caulobacter crescentus depends on the position of the gene in the chromosome
- PMID: 24891444
- PMCID: PMC4135674
- DOI: 10.1128/JB.01516-14
Localization of the outer membrane protein OmpA2 in Caulobacter crescentus depends on the position of the gene in the chromosome
Abstract
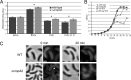
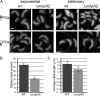
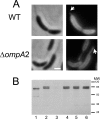
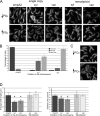
The outer membrane of Gram-negative bacteria is an essential structure involved in nutrient uptake, protection against harmful substances, and cell growth. Different proteins keep the outer membrane from blebbing out by simultaneously interacting with it and with the cell wall. These proteins have been mainly studied in enterobacteria, where OmpA and the Braun and Pal lipoproteins stabilize the outer membrane. Some degree of functional redundancy exists between these proteins, since none of them is essential but the absence of two of them results in a severe phenotype. Caulobacter crescentus has a different strategy to maintain its outer membrane, since it lacks the Braun lipoprotein and Pal is essential. In this work, we characterized OmpA2, an OmpA-like protein, in this bacterium. Our results showed that this protein is required for normal stalk growth and that it plays a minor role in the stability of the outer membrane. An OmpA2 fluorescent fusion protein showed that the concentration of this protein decreases from the stalk to the new pole. This localization pattern is important for its function, and it depends on the position of the gene locus in the chromosome and, as a consequence, in the cell. This result suggests that little diffusion occurs from the moment that the gene is transcribed until the mature protein attaches to the cell wall in the periplasm. This mechanism reveals the integration of different levels of information from protein function down to genome arrangement that allows the cell to self-organize.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures

References
-
- Braun V, Rehn K. 1969. Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. Eur. J. Biochem. 10:426–438 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

