Transcription start site sequence and spacing between the -10 region and the start site affect reiterative transcription-mediated regulation of gene expression in Escherichia coli
- PMID: 24891446
- PMCID: PMC4135645
- DOI: 10.1128/JB.01753-14
Transcription start site sequence and spacing between the -10 region and the start site affect reiterative transcription-mediated regulation of gene expression in Escherichia coli
Abstract
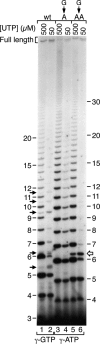
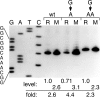
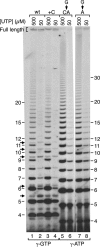
Reiterative transcription is a reaction catalyzed by RNA polymerase, in which nucleotides are repetitively added to the 3' end of a nascent transcript due to upstream slippage of the transcript without movement of the DNA template. In Escherichia coli, the expression of several operons is regulated through mechanisms in which high intracellular levels of UTP promote reiterative transcription that adds extra U residues to the 3' end of a nascent transcript during transcription initiation. Immediately following the addition of one or more extra U residues, the nascent transcripts are released from the transcription initiation complex, thereby reducing the level of gene expression. Therefore, gene expression can be regulated by internal UTP levels, which reflect the availability of external pyrimidine sources. The magnitude of gene regulation by these mechanisms varies considerably, even when control mechanisms are analogous. These variations apparently are due to differences in promoter sequences. One of the operons regulated (in part) by UTP-sensitive reiterative transcription in E. coli is the carAB operon, which encodes the first enzyme in the pyrimidine nucleotide biosynthetic pathway. In this study, we used the carAB operon to examine the effects of nucleotide sequence at and near the transcription start site and spacing between the start site and -10 region of the promoter on reiterative transcription and gene regulation. Our results indicate that these variables are important determinants in establishing the extent of reiterative transcription, levels of productive transcription, and range of gene regulation.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures



Similar articles
-
Regulation of codBA operon expression in Escherichia coli by UTP-dependent reiterative transcription and UTP-sensitive transcriptional start site switching.J Mol Biol. 1995 Dec 8;254(4):552-65. doi: 10.1006/jmbi.1995.0638. J Mol Biol. 1995. PMID: 7500333
-
Regulation of carAB expression in Escherichia coli occurs in part through UTP-sensitive reiterative transcription.J Bacteriol. 1998 Feb;180(3):705-13. doi: 10.1128/JB.180.3.705-713.1998. J Bacteriol. 1998. PMID: 9457878 Free PMC article.
-
Regulation of upp expression in Escherichia coli by UTP-sensitive selection of transcriptional start sites coupled with UTP-dependent reiterative transcription.J Bacteriol. 1997 Nov;179(21):6665-73. doi: 10.1128/jb.179.21.6665-6673.1997. J Bacteriol. 1997. PMID: 9352914 Free PMC article.
-
Regulation of pyrimidine biosynthetic gene expression in bacteria: repression without repressors.Microbiol Mol Biol Rev. 2008 Jun;72(2):266-300, table of contents. doi: 10.1128/MMBR.00001-08. Microbiol Mol Biol Rev. 2008. PMID: 18535147 Free PMC article. Review.
-
Regulation of gene expression by reiterative transcription.Curr Opin Microbiol. 2011 Apr;14(2):142-7. doi: 10.1016/j.mib.2011.01.012. Epub 2011 Feb 19. Curr Opin Microbiol. 2011. PMID: 21334966 Free PMC article. Review.
Cited by
-
Control of Transcription Initiation by Biased Thermal Fluctuations on Repetitive Genomic Sequences.Biomolecules. 2020 Sep 9;10(9):1299. doi: 10.3390/biom10091299. Biomolecules. 2020. PMID: 32916947 Free PMC article.
-
Natural C-independent expression of restriction endonuclease in a C protein-associated restriction-modification system.Nucleic Acids Res. 2016 Apr 7;44(6):2646-60. doi: 10.1093/nar/gkv1331. Epub 2015 Dec 9. Nucleic Acids Res. 2016. PMID: 26656489 Free PMC article.
-
The Role of Pyrophosphorolysis in the Initiation-to-Elongation Transition by E. coli RNA Polymerase.J Mol Biol. 2019 Jun 28;431(14):2528-2542. doi: 10.1016/j.jmb.2019.04.020. Epub 2019 Apr 26. J Mol Biol. 2019. PMID: 31029704 Free PMC article.
-
Regulation of carbamoylphosphate synthesis in Escherichia coli: an amazing metabolite at the crossroad of arginine and pyrimidine biosynthesis.Amino Acids. 2018 Dec;50(12):1647-1661. doi: 10.1007/s00726-018-2654-z. Epub 2018 Sep 20. Amino Acids. 2018. PMID: 30238253 Free PMC article. Review.
-
Open complex scrunching before nucleotide addition accounts for the unusual transcription start site of E. coli ribosomal RNA promoters.Proc Natl Acad Sci U S A. 2016 Mar 29;113(13):E1787-95. doi: 10.1073/pnas.1522159113. Epub 2016 Mar 14. Proc Natl Acad Sci U S A. 2016. PMID: 26976590 Free PMC article.
References
-
- Anikin M, Molodtsov V, Temiakov D, McAllister WT. 2010. Transcript slippage and recoding, p 409–432 In Atkins JF, Gesteland RF. (ed), Recoding: expansion of decoding rules enriches gene expression. Nucleic acids and molecular biology, vol 24 Springer, New York, NY
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

