Phosphorylation of tropomodulin1 contributes to the regulation of actin filament architecture in cardiac muscle
- PMID: 24891520
- PMCID: PMC4139905
- DOI: 10.1096/fj.13-246009
Phosphorylation of tropomodulin1 contributes to the regulation of actin filament architecture in cardiac muscle
Abstract
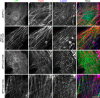
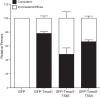
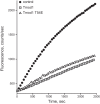
Tropomodulin1 (Tmod1) is an actin-capping protein that plays an important role in actin filament pointed-end dynamics and length in striated muscle. No mechanisms have been identified to explain how Tmod1's functional properties are regulated. The purpose of this investigation was to explore the functional significance of the phosphorylation of Tmod1 at previously identified Thr54. Rat cardiomyocytes were assessed for phosphorylation of Tmod1 using Pro-Q Diamond staining and (32)P labeling. Green fluorescent protein-tagged phosphorylation-mimic (T54E) and phosphorylation-deficient (T54A) versions of Tmod1 were expressed in cultured cardiomyocytes, and the ability of these mutants to assemble and restrict actin lengths was observed. We report for the first time that Tmod1 is phosphorylated endogenously in cardiomyocytes, and phosphorylation at Thr54 causes a significant reduction in the ability of Tmod1 to assemble to the pointed end compared with that of the wild type (WT; 48 vs. 78%, respectively). In addition, overexpression of Tmod1-T54E restricts actin filament lengths by only ∼3%, whereas Tmod1-WT restricts the lengths significantly by ∼8%. Finally, Tmod1-T54E altered the actin filament-capping activity in polymerization assays. Taken together, our data suggest that pointed-end assembly and Tmod1's thin filament length regulatory function are regulated by its phosphorylation state.
Keywords: post-translational modification; sarcomere.
© FASEB.
Figures



References
-
- Cox P. R., Zoghbi H. Y. (2000) Sequencing, expression analysis, and mapping of three unique human tropomodulin genes and their mouse orthologs. Genomics 63, 97–107 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

