Identification of unique SUN-interacting nuclear envelope proteins with diverse functions in plants
- PMID: 24891605
- PMCID: PMC4050730
- DOI: 10.1083/jcb.201401138
Identification of unique SUN-interacting nuclear envelope proteins with diverse functions in plants
Abstract
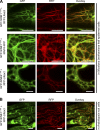
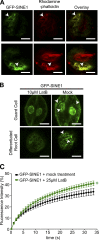
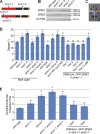
Although a plethora of nuclear envelope (NE) transmembrane proteins (NETs) have been identified in opisthokonts, plant NETs are largely unknown. The only known NET homologues in plants are Sad1/UNC-84 (SUN) proteins, which bind Klarsicht/ANC-1/Syne-1 homology (KASH) proteins. Therefore, de novo identification of plant NETs is necessary. Based on similarities between opisthokont KASH proteins and the only known plant KASH proteins, WPP domain-interacting proteins, we used a computational method to identify the KASH subset of plant NETs. Ten potential plant KASH protein families were identified, and five candidates from four of these families were verified for their NE localization, depending on SUN domain interaction. Of those, Arabidopsis thaliana SINE1 is involved in actin-dependent nuclear positioning in guard cells, whereas its paralogue SINE2 contributes to innate immunity against an oomycete pathogen. This study dramatically expands our knowledge of plant KASH proteins and suggests that plants and opisthokonts have recruited different KASH proteins to perform NE regulatory functions.
© 2014 Zhou et al.
Figures



References
-
- Caillaud M.-C., Piquerez S.J.M., Fabro G., Steinbrenner J., Ishaque N., Beynon J., Jones J.D.G. 2012. Subcellular localization of the Hpa RxLR effector repertoire identifies a tonoplast-associated protein HaRxL17 that confers enhanced plant susceptibility. Plant J. 69:252–265 10.1111/j.1365-313X.2011.04787.x - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

