RNA-binding protein AUF1 promotes myogenesis by regulating MEF2C expression levels
- PMID: 24891619
- PMCID: PMC4135590
- DOI: 10.1128/MCB.00423-14
RNA-binding protein AUF1 promotes myogenesis by regulating MEF2C expression levels
Abstract
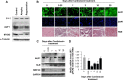
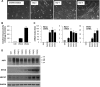
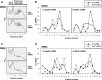
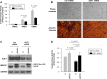
The mammalian RNA-binding protein AUF1 (AU-binding factor 1, also known as heterogeneous nuclear ribonucleoprotein D [hnRNP D]) binds to numerous mRNAs and influences their posttranscriptional fate. Given that many AUF1 target mRNAs encode muscle-specific factors, we investigated the function of AUF1 in skeletal muscle differentiation. In mouse C2C12 myocytes, where AUF1 levels rise at the onset of myogenesis and remain elevated throughout myocyte differentiation into myotubes, RNP immunoprecipitation (RIP) analysis indicated that AUF1 binds prominently to Mef2c (myocyte enhancer factor 2c) mRNA, which encodes the key myogenic transcription factor MEF2C. By performing mRNA half-life measurements and polysome distribution analysis, we found that AUF1 associated with the 3' untranslated region (UTR) of Mef2c mRNA and promoted MEF2C translation without affecting Mef2c mRNA stability. In addition, AUF1 promoted Mef2c gene transcription via a lesser-known role of AUF1 in transcriptional regulation. Importantly, lowering AUF1 delayed myogenesis, while ectopically restoring MEF2C expression levels partially rescued the impairment of myogenesis seen after reducing AUF1 levels. We propose that MEF2C is a key effector of the myogenesis program promoted by AUF1.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
