The PTEN/NRF2 axis promotes human carcinogenesis
- PMID: 24892215
- PMCID: PMC4245871
- DOI: 10.1089/ars.2014.5843
The PTEN/NRF2 axis promotes human carcinogenesis
Abstract
Aims: A recent study conducted in mice reported that liver-specific knockout of tumor suppressor Pten augments nuclear factor (erythroid-derived 2)-like 2 (NRF2) transcriptional activity. Here, we further investigated how phosphatase and tensin homolog deleted on chromosome 10 (PTEN) controls NRF2 and the relevance of this pathway in human carcin ogenesis.
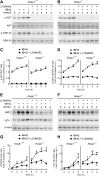
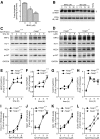
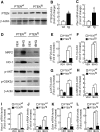
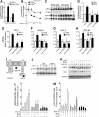
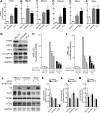
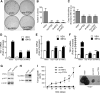
Results: Drug and genetic targeting to PTEN and phosphoproteomics approaches indicated that PTEN leads to glycogen synthase kinase-3 (GSK-3)-mediated phosphorylation of NRF2 at residues Ser(335) and Ser(338) and subsequent beta-transducin repeat containing protein (β-TrCP)-dependent but Kelch-like ECH-associated protein 1 (KEAP1)-independent degradation. Rescue experiments in PTEN-deficient cells and xerographs in athymic mice indicated that loss of PTEN leads to increased NRF2 signature which provides a proliferating and tumorigenic advantage. Tissue microarrays from endometrioid carcinomas showed that 80% of PTEN-negative tumors expressed high levels of NRF2 or its target heme oxygenase-1 (HO-1).
Innovation: These results uncover a new mechanism of oncogenic activation of NRF2 by loss of its negative regulation by PTEN/GSK-3/β-TrCP that may be relevant to a large number of tumors, including endometrioid carcinomas.
Conclusion: Increased activity of NRF2 due to loss of PTEN is instrumental in human carcinogenesis and represents a novel therapeutic target.
Figures


Similar articles
-
Nrf2 enhances cholangiocyte expansion in Pten-deficient livers.Mol Cell Biol. 2014 Mar;34(5):900-13. doi: 10.1128/MCB.01384-13. Epub 2013 Dec 30. Mol Cell Biol. 2014. PMID: 24379438 Free PMC article.
-
Guggulsterone induces heme oxygenase-1 expression through activation of Nrf2 in human mammary epithelial cells: PTEN as a putative target.Carcinogenesis. 2012 Feb;33(2):368-76. doi: 10.1093/carcin/bgr259. Epub 2011 Nov 17. Carcinogenesis. 2012. PMID: 22095074
-
Estrogen increases Nrf2 activity through activation of the PI3K pathway in MCF-7 breast cancer cells.Exp Cell Res. 2014 Nov 1;328(2):351-60. doi: 10.1016/j.yexcr.2014.08.030. Epub 2014 Aug 27. Exp Cell Res. 2014. PMID: 25172557
-
Dual regulation of transcription factor Nrf2 by Keap1 and by the combined actions of β-TrCP and GSK-3.Biochem Soc Trans. 2015 Aug;43(4):611-20. doi: 10.1042/BST20150011. Epub 2015 Aug 3. Biochem Soc Trans. 2015. PMID: 26551701 Review.
-
Structural and functional characterization of Nrf2 degradation by glycogen synthase kinase 3/β-TrCP.Free Radic Biol Med. 2015 Nov;88(Pt B):147-157. doi: 10.1016/j.freeradbiomed.2015.04.029. Epub 2015 Apr 30. Free Radic Biol Med. 2015. PMID: 25937177 Review.
Cited by
-
VEGFA and NFE2L2 Gene Expression and Regulation by MicroRNAs in Thyroid Papillary Cancer and Colloid Goiter.Genes (Basel). 2020 Aug 19;11(9):954. doi: 10.3390/genes11090954. Genes (Basel). 2020. PMID: 32824922 Free PMC article.
-
Clinical Relevance of Biomarkers of Oxidative Stress.Antioxid Redox Signal. 2015 Nov 10;23(14):1144-70. doi: 10.1089/ars.2015.6317. Epub 2015 Oct 26. Antioxid Redox Signal. 2015. PMID: 26415143 Free PMC article. Review.
-
Redox regulation of the insulin signalling pathway.Redox Biol. 2021 Jun;42:101964. doi: 10.1016/j.redox.2021.101964. Epub 2021 Apr 2. Redox Biol. 2021. PMID: 33893069 Free PMC article. Review.
-
The redox biology network in cancer pathophysiology and therapeutics.Redox Biol. 2015 Aug;5:347-357. doi: 10.1016/j.redox.2015.06.014. Epub 2015 Jun 25. Redox Biol. 2015. PMID: 26122399 Free PMC article. Review.
-
The molecular biology and therapeutic potential of Nrf2 in leukemia.Cancer Cell Int. 2022 Jul 29;22(1):241. doi: 10.1186/s12935-022-02660-5. Cancer Cell Int. 2022. PMID: 35906617 Free PMC article. Review.
References
-
- Almazari I, Park JM, Park SA, Suh JY, Na HK, Cha YN, Surh YJ. Guggulsterone induces heme oxygenase-1 expression through activation of Nrf2 in human mammary epithelial cells: PTEN as a putative target. Carcinogenesis 33: 368–376, 2012 - PubMed
-
- DeNicola GM, Karreth FA, Humpton TJ, Gopinathan A, Wei C, Frese K, Mangal D, Yu KH, Yeo CJ, Calhoun ES, Scrimieri F, Winter JM, Hruban RH, Iacobuzio-Donahue C, Kern SE, Blair IA, and Tuveson DA. Oncogene-induced Nrf2 transcription promotes ROS detoxification and tumorigenesis. Nature 475: 106–109, 2011 - PMC - PubMed
-
- Dinkova-Kostova AT, and Wang XJ. Induction of the Keap1/Nrf2/ARE pathway by oxidizable diphenols. Chem Biol Interact 192: 101–106, 2011 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

