Abnormal B cell memory subsets dominate HIV-specific responses in infected individuals
- PMID: 24892810
- PMCID: PMC4071400
- DOI: 10.1172/JCI74351
Abnormal B cell memory subsets dominate HIV-specific responses in infected individuals
Abstract
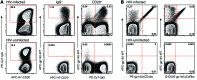
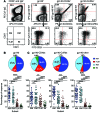
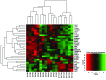
Recently, several neutralizing anti-HIV antibodies have been isolated from memory B cells of HIV-infected individuals. Despite extensive evidence of B cell dysfunction in HIV disease, little is known about the cells from which these rare HIV-specific antibodies originate. Accordingly, we used HIV envelope gp140 and CD4 or coreceptor (CoR) binding site (bs) mutant probes to evaluate HIV-specific responses in peripheral blood B cells of HIV-infected individuals at various stages of infection. In contrast to non-HIV responses, HIV-specific responses against gp140 were enriched within abnormal B cells, namely activated and exhausted memory subsets, which are largely absent in the blood of uninfected individuals. Responses against the CoRbs, which is a poorly neutralizing epitope, arose early, whereas those against the well-characterized neutralizing epitope CD4bs were delayed and infrequent. Enrichment of the HIV-specific response within resting memory B cells, the predominant subset in uninfected individuals, did occur in certain infected individuals who maintained low levels of plasma viremia and immune activation with or without antiretroviral therapy. The distribution of HIV-specific responses among memory B cell subsets was corroborated by transcriptional analyses. Taken together, our findings provide valuable insight into virus-specific B cell responses in HIV infection and demonstrate that memory B cell abnormalities may contribute to the ineffectiveness of the antibody response in infected individuals.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

