Towards a molecular characterization of autism spectrum disorders: an exome sequencing and systems approach
- PMID: 24893065
- PMCID: PMC4080319
- DOI: 10.1038/tp.2014.38
Towards a molecular characterization of autism spectrum disorders: an exome sequencing and systems approach
Abstract
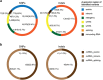
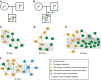
The hypothetical 'AXAS' gene network model that profiles functional patterns of heterogeneous DNA variants overrepresented in autism spectrum disorder (ASD), X-linked intellectual disability, attention deficit and hyperactivity disorder and schizophrenia was used in this current study to analyze whole exome sequencing data from an Australian ASD cohort. An optimized DNA variant filtering pipeline was used to identify loss-of-function DNA variations. Inherited variants from parents with a broader autism phenotype and de novo variants were found to be significantly associated with ASD. Gene ontology analysis revealed that putative rare causal variants cluster in key neurobiological processes and are overrepresented in functions involving neuronal development, signal transduction and synapse development including the neurexin trans-synaptic complex. We also show how a complex gene network model can be used to fine map combinations of inherited and de novo variations in families with ASD that converge in the L1CAM pathway. Our results provide an important step forward in the molecular characterization of ASD with potential for developing a tool to analyze the pathogenesis of individual affected families.
Figures


References
-
- Cristino AS, Williams SM, Hawi Z, An JY, Bellgrove MA, Schwartz CE, et al. Neurodevelopmental and neuropsychiatric disorders represent an interconnected molecular system. Mol Psychiatry. 2014;19:294–301. - PubMed
-
- Autism and Developmental Disabilities Monitoring Network Surveillance Year 2008 Principal Investigators, Centers for Disease Control and Prevention Prevalence of autism spectrum disorders—Autism and Developmental Disabilities Monitoring Network, 14 sites, United States, 2008. MMWR Surveill Summ. 2012;61:1–19. - PubMed
-
- Devlin B, Scherer SW. Genetic architecture in autism spectrum disorder. Curr Opin Genet Dev. 2012;22:229–237. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

