Metabolomic profiling and genomic study of a marine sponge-associated Streptomyces sp
- PMID: 24893324
- PMCID: PMC4071579
- DOI: 10.3390/md12063323
Metabolomic profiling and genomic study of a marine sponge-associated Streptomyces sp
Abstract
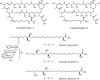
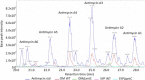
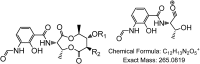
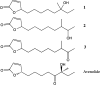
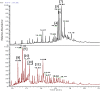
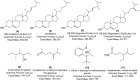
Metabolomics and genomics are two complementary platforms for analyzing an organism as they provide information on the phenotype and genotype, respectively. These two techniques were applied in the dereplication and identification of bioactive compounds from a Streptomyces sp. (SM8) isolated from the sponge Haliclona simulans from Irish waters. Streptomyces strain SM8 extracts showed antibacterial and antifungal activity. NMR analysis of the active fractions proved that hydroxylated saturated fatty acids were the major components present in the antibacterial fractions. Antimycin compounds were initially putatively identified in the antifungal fractions using LC-Orbitrap. Their presence was later confirmed by comparison to a standard. Genomic analysis of Streptomyces sp. SM8 revealed the presence of multiple secondary metabolism gene clusters, including a gene cluster for the biosynthesis of the antifungal antimycin family of compounds. The antimycin gene cluster of Streptomyces sp. SM8 was inactivated by disruption of the antimycin biosynthesis gene antC. Extracts from this mutant strain showed loss of antimycin production and significantly less antifungal activity than the wild-type strain. Three butenolides, 4,10-dihydroxy-10-methyl-dodec-2-en-1,4-olide (1), 4,11-dihydroxy-10-methyl-dodec-2-en-1,4-olide (2), and 4-hydroxy-10-methyl-11-oxo-dodec-2-en-1,4-olide (3) that had previously been reported from marine Streptomyces species were also isolated from SM8. Comparison of the extracts of Streptomyces strain SM8 and its host sponge, H. simulans, using LC-Orbitrap revealed the presence of metabolites common to both extracts, providing direct evidence linking sponge metabolites to a specific microbial symbiont.
Figures






Similar articles
-
Draft Genome Sequence of the Antimycin-Producing Bacterium Streptomyces sp. Strain SM8, Isolated from the Marine Sponge Haliclona simulans.Genome Announc. 2018 Jan 25;6(4):e01535-17. doi: 10.1128/genomeA.01535-17. Genome Announc. 2018. PMID: 29371360 Free PMC article.
-
Streptomyces associated with a marine sponge Haliclona sp.; biosynthetic genes for secondary metabolites and products.Environ Microbiol. 2011 Feb;13(2):391-403. doi: 10.1111/j.1462-2920.2010.02337.x. Epub 2010 Sep 16. Environ Microbiol. 2011. PMID: 20849448
-
Purification and biological evaluation of the metabolites produced by Streptomyces sp. TK-VL_333.Res Microbiol. 2010 Jun;161(5):335-45. doi: 10.1016/j.resmic.2010.03.011. Epub 2010 Apr 18. Res Microbiol. 2010. PMID: 20403429
-
Multi-omic investigation identifies key antifungal biochemistry during fermentation of a Streptomyces biological control agent.Microbiol Res. 2025 Mar;292:128032. doi: 10.1016/j.micres.2024.128032. Epub 2024 Dec 18. Microbiol Res. 2025. PMID: 39721340
-
N-Acetyl-Cysteinylated Streptophenazines from Streptomyces.J Nat Prod. 2022 May 27;85(5):1239-1247. doi: 10.1021/acs.jnatprod.1c01123. Epub 2022 Apr 14. J Nat Prod. 2022. PMID: 35422124 Free PMC article. Review.
Cited by
-
Marine Natural Products: A Source of Novel Anticancer Drugs.Mar Drugs. 2019 Aug 23;17(9):491. doi: 10.3390/md17090491. Mar Drugs. 2019. PMID: 31443597 Free PMC article. Review.
-
Regulation of Antibiotic Production by Signaling Molecules in Streptomyces.Front Microbiol. 2019 Dec 19;10:2927. doi: 10.3389/fmicb.2019.02927. eCollection 2019. Front Microbiol. 2019. PMID: 31921086 Free PMC article. Review.
-
Defining the biomarkers in anti-MRSA fractions of soil Streptomycetes by multivariate analysis.Antonie Van Leeuwenhoek. 2025 May 10;118(6):77. doi: 10.1007/s10482-025-02087-8. Antonie Van Leeuwenhoek. 2025. PMID: 40347296
-
Butenolides from Streptomyces albus J1074 Act as External Signals To Stimulate Avermectin Production in Streptomyces avermitilis.Appl Environ Microbiol. 2018 Apr 16;84(9):e02791-17. doi: 10.1128/AEM.02791-17. Print 2018 May 1. Appl Environ Microbiol. 2018. PMID: 29500256 Free PMC article.
-
Draft Genome Sequence of the Antimycin-Producing Bacterium Streptomyces sp. Strain SM8, Isolated from the Marine Sponge Haliclona simulans.Genome Announc. 2018 Jan 25;6(4):e01535-17. doi: 10.1128/genomeA.01535-17. Genome Announc. 2018. PMID: 29371360 Free PMC article.
References
-
- Schmitz F., Vanderah D., Hollenbeak K., Enwall C., Gopichand Y., Sengupta P., Hossain M., Vanderhelm D. Metabolites from the marine sponge Tedania ignis. A new atisanediol and several known diketopiperazines. J. Org. Chem. 1983;48:3941–3945. doi: 10.1021/jo00170a011. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

