Overexpression of GmAKT2 potassium channel enhances resistance to soybean mosaic virus
- PMID: 24893844
- PMCID: PMC4074861
- DOI: 10.1186/1471-2229-14-154
Overexpression of GmAKT2 potassium channel enhances resistance to soybean mosaic virus
Abstract
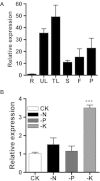
Background: Soybean mosaic virus (SMV) is the most prevalent viral disease in many soybean production areas. Due to a large number of SMV resistant loci and alleles, SMV strains and the rapid evolution in avirulence/effector genes, traditional breeding for SMV resistance is complex. Genetic engineering is an effective alternative method for improving SMV resistance in soybean. Potassium (K+) is the most abundant inorganic solute in plant cells, and is involved in plant responses to abiotic and biotic stresses. Studies have shown that altering the level of K+ status can reduce the spread of the viral diseases. Thus K+ transporters are putative candidates to target for soybean virus resistance.
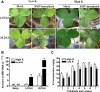
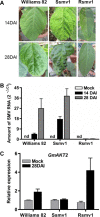
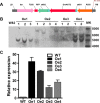
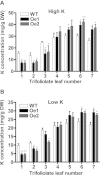
Results: The addition of K+ fertilizer significantly reduced SMV incidence. Analysis of K+ channel gene expression indicated that GmAKT2, the ortholog of Arabidopsis K+ weak channel encoding gene AKT2, was significantly induced by SMV inoculation in the SMV highly-resistant genotype Rsmv1, but not in the susceptible genotype Ssmv1. Transgenic soybean plants overexpressing GmAKT2 were produced and verified by Southern blot and RT-PCR analysis. Analysis of K+ concentrations on different leaves of both the transgenic and the wildtype (Williams 82) plants revealed that overexpression of GmAKT2 significantly increased K+ concentrations in young leaves of plants. In contrast, K+ concentrations in the old leaves of the GmAKT2-Oe plants were significantly lower than those in WT plants. These results indicated that GmAKT2 acted as a K+ transporter and affected the distribution of K+ in soybean plants. Starting from 14 days after inoculation (DAI) of SMV G7, severe mosaic symptoms were observed on the WT leaves. In contrast, the GmAKT2-Oe plants showed no symptom of SMV infection. At 14 and 28 DAI, the amount of SMV RNA in WT plants increased 200- and 260- fold relative to GmAKT2-Oe plants at each time point. Thus, SMV development was significantly retarded in GmAKT2-overexpressing transgenic soybean plants.
Conclusions: Overexpression of GmAKT2 significantly enhanced SMV resistance in transgenic soybean. Thus, alteration of K+ transporter expression is a novel molecular approach for enhancing SMV resistance in soybean.
Figures

Similar articles
-
Over-expression of GmSN1 enhances virus resistance in Arabidopsis and soybean.Plant Cell Rep. 2017 Sep;36(9):1441-1455. doi: 10.1007/s00299-017-2167-3. Epub 2017 Jun 27. Plant Cell Rep. 2017. PMID: 28656325
-
Characterization of Soybean mosaic virus resistance derived from inverted repeat-SMV-HC-Pro genes in multiple soybean cultivars.Theor Appl Genet. 2015 Aug;128(8):1489-505. doi: 10.1007/s00122-015-2522-0. Epub 2015 May 1. Theor Appl Genet. 2015. PMID: 25930057
-
Overexpression of a GmCnx1 gene enhanced activity of nitrate reductase and aldehyde oxidase, and boosted mosaic virus resistance in soybean.PLoS One. 2015 Apr 17;10(4):e0124273. doi: 10.1371/journal.pone.0124273. eCollection 2015. PLoS One. 2015. PMID: 25886067 Free PMC article.
-
Decades of Genetic Research on Soybean mosaic virus Resistance in Soybean.Viruses. 2022 May 24;14(6):1122. doi: 10.3390/v14061122. Viruses. 2022. PMID: 35746594 Free PMC article. Review.
-
Overexpression of an aquaporin protein from Aspergillus glaucus confers salt tolerance in transgenic soybean.Transgenic Res. 2021 Dec;30(6):727-737. doi: 10.1007/s11248-021-00280-9. Epub 2021 Aug 30. Transgenic Res. 2021. PMID: 34460070 Review.
Cited by
-
Over-expression of GmKR3, a TIR-NBS-LRR type R gene, confers resistance to multiple viruses in soybean.Plant Mol Biol. 2019 Jan;99(1-2):95-111. doi: 10.1007/s11103-018-0804-z. Epub 2018 Dec 10. Plant Mol Biol. 2019. PMID: 30535849
-
Genome-Wide Association Studies of Root-Related Traits in Brassica napus L. under Low-Potassium Conditions.Plants (Basel). 2022 Jul 12;11(14):1826. doi: 10.3390/plants11141826. Plants (Basel). 2022. PMID: 35890461 Free PMC article.
-
A comprehensive review on Gossypium hirsutum resistance against cotton leaf curl virus.Front Genet. 2024 Feb 19;15:1306469. doi: 10.3389/fgene.2024.1306469. eCollection 2024. Front Genet. 2024. PMID: 38440193 Free PMC article. Review.
-
The Current Status of the Soybean-Soybean Mosaic Virus (SMV) Pathosystem.Front Microbiol. 2016 Nov 30;7:1906. doi: 10.3389/fmicb.2016.01906. eCollection 2016. Front Microbiol. 2016. PMID: 27965641 Free PMC article. Review.
-
Global transcriptome changes in perennial ryegrass during early infection by pink snow mould.Sci Rep. 2016 Jun 27;6:28702. doi: 10.1038/srep28702. Sci Rep. 2016. PMID: 27346054 Free PMC article.
References
-
- Cui X, Chen X, Wang A. In: Soybean - Molecular Aspects of Breeding. Sudaric A, editor. Shanghai: InTech; 2011. Detection, understanding and control of soybean mosaic virus; pp. 335–354.
-
- Zheng C, Chen P, Gergerich R. Effect of temperature on the expression of necrosis in soybean infected with soybean mosaic virus. Crop Sci. 2005;45(3):916–922. doi: 10.2135/cropsci2004.0286. - DOI
-
- Hill JH. In: Compendium of Soybean Diseases. 4. Hartman GL, Sinclair JB, Rupe JC, editor. St Paul, MN: American Phytopathological Society; 1999. Soybean mosaic virus; pp. 70–71.
-
- Buzzell RI, Tu JC. Inheritance of a soybean stem-tip necrosis reaction to soybean mosaic virus. J Hered. 1989;80(5):400–401.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

