A Snail1/Notch1 signalling axis controls embryonic vascular development
- PMID: 24894949
- PMCID: PMC4052376
- DOI: 10.1038/ncomms4998
A Snail1/Notch1 signalling axis controls embryonic vascular development
Abstract
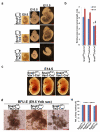
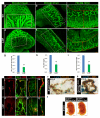
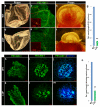
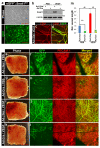
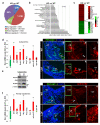
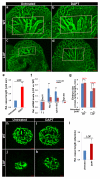
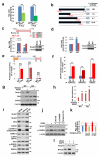
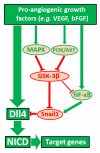
Notch1-Delta-like 4 (Dll4) signalling controls vascular development by regulating endothelial cell (EC) targets that modulate vessel wall remodelling and arterial-venous specification. The molecular effectors that modulate Notch signalling during vascular development remain largely undefined. Here we demonstrate that the transcriptional repressor, Snail1, acts as a VEGF-induced regulator of Notch1 signalling and Dll4 expression. EC-specific Snail1 loss-of-function conditional knockout mice die in utero with defects in vessel wall remodelling in association with losses in mural cell investment and disruptions in arterial-venous specification. Snail1 loss-of-function conditional knockout embryos further display upregulated Notch1 signalling and Dll4 expression that is partially reversed by inhibiting γ-secretase activity in vivo with Dll4 identified as a direct target of Snail1-mediated transcriptional repression. These results document a Snail1-Dll4/Notch1 axis that controls embryonic vascular development.
Figures
References
-
- Phng LK, Gerhardt H. Angiogenesis: a team effort coordinated by Notch. Dev. Cell. 2009;16:196–208. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

