Poly(ADP-ribose) polymerase inhibitors sensitize cancer cells to death receptor-mediated apoptosis by enhancing death receptor expression
- PMID: 24895135
- PMCID: PMC4110268
- DOI: 10.1074/jbc.M114.549220
Poly(ADP-ribose) polymerase inhibitors sensitize cancer cells to death receptor-mediated apoptosis by enhancing death receptor expression
Abstract
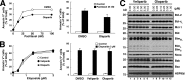
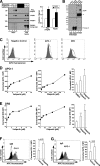
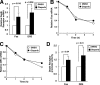
Recombinant human tumor necrosis factor-α-related apoptosis inducing ligand (TRAIL), agonistic monoclonal antibodies to TRAIL receptors, and small molecule TRAIL receptor agonists are in various stages of preclinical and early phase clinical testing as potential anticancer drugs. Accordingly, there is substantial interest in understanding factors that affect sensitivity to these agents. In the present study we observed that the poly(ADP-ribose) polymerase (PARP) inhibitors olaparib and veliparib sensitize the myeloid leukemia cell lines ML-1 and K562, the ovarian cancer line PEO1, non-small cell lung cancer line A549, and a majority of clinical AML isolates, but not normal marrow, to TRAIL. Further analysis demonstrated that PARP inhibitor treatment results in activation of the FAS and TNFRSF10B (death receptor 5 (DR5)) promoters, increased Fas and DR5 mRNA, and elevated cell surface expression of these receptors in sensitized cells. Chromatin immunoprecipitation demonstrated enhanced binding of the transcription factor Sp1 to the TNFRSF10B promoter in the presence of PARP inhibitor. Knockdown of PARP1 or PARP2 (but not PARP3 and PARP4) not only increased expression of Fas and DR5 at the mRNA and protein level, but also recapitulated the sensitizing effects of the PARP inhibition. Conversely, Sp1 knockdown diminished the PARP inhibitor effects. In view of the fact that TRAIL is part of the armamentarium of natural killer cells, these observations identify a new facet of PARP inhibitor action while simultaneously providing the mechanistic underpinnings of a novel therapeutic combination that warrants further investigation.
Figures



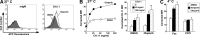


Similar articles
-
PARP inhibition restores extrinsic apoptotic sensitivity in glioblastoma.PLoS One. 2014 Dec 22;9(12):e114583. doi: 10.1371/journal.pone.0114583. eCollection 2014. PLoS One. 2014. PMID: 25531448 Free PMC article.
-
The poly(ADP-ribose) polymerase inhibitor olaparib induces up-regulation of death receptors in primary acute myeloid leukemia blasts by NF-κB activation.Cancer Lett. 2018 Jun 1;423:127-138. doi: 10.1016/j.canlet.2018.03.008. Epub 2018 Mar 8. Cancer Lett. 2018. PMID: 29526802
-
PARP-1 regulates resistance of pancreatic cancer to TRAIL therapy.Clin Cancer Res. 2013 Sep 1;19(17):4750-9. doi: 10.1158/1078-0432.CCR-13-0516. Epub 2013 Jul 5. Clin Cancer Res. 2013. PMID: 23833311 Free PMC article.
-
Regulation of TRAIL-receptor expression by the ubiquitin-proteasome system.Int J Mol Sci. 2014 Oct 14;15(10):18557-73. doi: 10.3390/ijms151018557. Int J Mol Sci. 2014. PMID: 25318057 Free PMC article. Review.
-
Review of poly (ADP-ribose) polymerase (PARP) mechanisms of action and rationale for targeting in cancer and other diseases.Crit Rev Eukaryot Gene Expr. 2014;24(1):15-28. doi: 10.1615/critreveukaryotgeneexpr.2013006875. Crit Rev Eukaryot Gene Expr. 2014. PMID: 24579667 Free PMC article. Review.
Cited by
-
Candidate Markers of Olaparib Response from Genomic Data Analyses of Human Cancer Cell Lines.Cancers (Basel). 2021 Mar 15;13(6):1296. doi: 10.3390/cancers13061296. Cancers (Basel). 2021. PMID: 33803939 Free PMC article.
-
The cAMP analogs have potent anti-proliferative effects on medullary thyroid cancer cell lines.Endocrine. 2016 Jan;51(1):101-12. doi: 10.1007/s12020-015-0597-7. Epub 2015 Apr 12. Endocrine. 2016. PMID: 25863490
-
Role of BRCA Mutations in Cancer Treatment with Poly(ADP-ribose) Polymerase (PARP) Inhibitors.Cancers (Basel). 2018 Dec 4;10(12):487. doi: 10.3390/cancers10120487. Cancers (Basel). 2018. PMID: 30518089 Free PMC article. Review.
-
A Phase I Clinical Trial of the Poly(ADP-ribose) Polymerase Inhibitor Veliparib and Weekly Topotecan in Patients with Solid Tumors.Clin Cancer Res. 2018 Feb 15;24(4):744-752. doi: 10.1158/1078-0432.CCR-17-1590. Epub 2017 Nov 14. Clin Cancer Res. 2018. PMID: 29138343 Free PMC article. Clinical Trial.
-
Olaparib Synergizes the Anticancer Activity of Daunorubicin via Interaction with AKR1C3.Cancers (Basel). 2020 Oct 26;12(11):3127. doi: 10.3390/cancers12113127. Cancers (Basel). 2020. PMID: 33114555 Free PMC article.
References
-
- Siegel R., Naishadham D., Jemal A. (2012) Cancer statistics, 2012. CA Cancer J. Clin. 62, 10–29 - PubMed
-
- Ozören N., El-Deiry W. S. (2003) Cell surface Death Receptor signaling in normal and cancer cells. Semin. Cancer Biol. 13, 135–147 - PubMed
-
- Yang A., Wilson N. S., Ashkenazi A. (2010) Proapoptotic DR4 and DR5 signaling in cancer cells: toward clinical translation. Curr. Opin. Cell Biol. 22, 837–844 - PubMed
-
- Wilson N. S., Yang A., Yang B., Couto S., Stern H., Gogineni A., Pitti R., Marsters S., Weimer R. M., Singh M., Ashkenazi A. (2012) Proapoptotic activation of death receptor 5 on tumor endothelial cells disrupts the vasculature and reduces tumor growth. Cancer Cell 22, 80–90 - PubMed
-
- Taylor R. C., Cullen S. P., Martin S. J. (2008) Apoptosis: controlled demolition at the cellular level. Nat. Rev. Mol. Cell biology 9, 231–241 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

