Invasion of the central nervous system by Cryptococcus neoformans requires a secreted fungal metalloprotease
- PMID: 24895304
- PMCID: PMC4049100
- DOI: 10.1128/mBio.01101-14
Invasion of the central nervous system by Cryptococcus neoformans requires a secreted fungal metalloprotease
Abstract
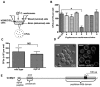
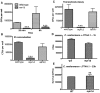
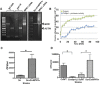
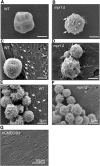
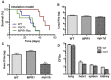
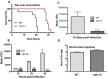
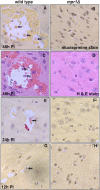
Cryptococcus spp. cause life-threatening fungal infection of the central nervous system (CNS), predominantly in patients with a compromised immune system. Why Cryptococcus neoformans has this remarkable tropism for the CNS is not clear. Recent research on cerebral pathogenesis of C. neoformans revealed a predominantly transcellular migration of cryptococci across the brain endothelium; however, the identities of key fungal virulence factors that function specifically to invade the CNS remain unresolved. Here we found that a novel, secreted metalloprotease (Mpr1) that we identified in the extracellular proteome of C. neoformans (CnMpr1) is required for establishing fungal disease in the CNS. Mpr1 belongs to a poorly characterized M36 class of fungalysins that are expressed in only some fungal species. A strain of C. neoformans lacking the gene encoding Mpr1 (mpr1Δ) failed to breach the endothelium in an in vitro model of the human blood-brain barrier (BBB). A mammalian host infected with the mpr1Δ null strain demonstrated significant improvement in survival due to a reduced brain fungal burden and lacked the brain pathology commonly associated with cryptococcal disease. The in vivo studies further indicate that Mpr1 is not required for fungal dissemination and Mpr1 likely targets the brain endothelium specifically. Remarkably, the sole expression of CnMPR1 in Saccharomyces cerevisiae resulted in a robust migration of yeast cells across the brain endothelium, demonstrating Mpr1's specific activity in breaching the BBB and suggesting that Mpr1 may function independently of the hyaluronic acid-CD44 pathway. This distinct role for Mpr1 may develop into innovative treatment options and facilitate a brain-specific drug delivery platform.
Importance: Cryptococcus neoformans is a medically relevant fungal pathogen causing significant morbidity and mortality, particularly in immunocompromised individuals. An intriguing feature is its strong neurotropism, and consequently the hallmark of cryptococcal disease is a brain infection, cryptococcal meningoencephalitis. For C. neoformans to penetrate the central nervous system (CNS), it first breaches the blood-brain barrier via a transcellular pathway; however, the identities of fungal factors required for this transmigration remain largely unknown. In an effort to identify extracellular fungal proteins that could mediate interactions with the brain endothelium, we undertook a proteomic analysis of the extracellular proteome and identified a secreted metalloprotease (Mpr1) belonging to the M36 class of fungalysins. Here we found that Mpr1 promotes migration of C. neoformans across the brain endothelium and into the CNS by facilitating attachment of cryptococci to the endothelium surface, thus underscoring the critical role of M36 proteases in fungal pathogenesis.
Copyright © 2014 Vu et al.
Figures
Similar articles
-
The Metalloprotease, Mpr1, Engages AnnexinA2 to Promote the Transcytosis of Fungal Cells across the Blood-Brain Barrier.Front Cell Infect Microbiol. 2017 Jun 30;7:296. doi: 10.3389/fcimb.2017.00296. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28713781 Free PMC article.
-
The structure-function analysis of the Mpr1 metalloprotease determinants of activity during migration of fungal cells across the blood-brain barrier.PLoS One. 2018 Aug 30;13(8):e0203020. doi: 10.1371/journal.pone.0203020. eCollection 2018. PLoS One. 2018. PMID: 30161190 Free PMC article.
-
An Antivirulence Approach for Preventing Cryptococcus neoformans from Crossing the Blood-Brain Barrier via Novel Natural Product Inhibitors of a Fungal Metalloprotease.mBio. 2020 Jul 21;11(4):e01249-20. doi: 10.1128/mBio.01249-20. mBio. 2020. PMID: 32694141 Free PMC article.
-
[Mechanism of Cryptococcus Meningoencephalitis].Med Mycol J. 2016;57(1):J27-32. doi: 10.3314/mmj.57.J27. Med Mycol J. 2016. PMID: 26936349 Review. Japanese.
-
Cryptococcal pathogenic mechanisms: a dangerous trip from the environment to the brain.Mem Inst Oswaldo Cruz. 2018;113(7):e180057. doi: 10.1590/0074-02760180057. Epub 2018 Apr 16. Mem Inst Oswaldo Cruz. 2018. PMID: 29668825 Free PMC article. Review.
Cited by
-
Disseminated Cryptococcus over pancreas, lung, and brain: a case report.J Med Case Rep. 2024 Oct 23;18(1):513. doi: 10.1186/s13256-024-04836-1. J Med Case Rep. 2024. PMID: 39438983 Free PMC article.
-
Fungal Infection in the Brain: What We Learned from Intravital Imaging.Front Immunol. 2016 Aug 2;7:292. doi: 10.3389/fimmu.2016.00292. eCollection 2016. Front Immunol. 2016. PMID: 27532000 Free PMC article. Review.
-
Transcriptomic Analysis of Extracellular RNA Governed by the Endocytic Adaptor Protein Cin1 of Cryptococcus deneoformans.Front Cell Infect Microbiol. 2020 Jun 23;10:256. doi: 10.3389/fcimb.2020.00256. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 32656093 Free PMC article.
-
Dancing cheek to cheek: Cryptococcus neoformans and phagocytes.Springerplus. 2015 Aug 12;4:410. doi: 10.1186/s40064-015-1192-3. eCollection 2015. Springerplus. 2015. PMID: 26266081 Free PMC article. Review.
-
New strategies for combating fungal infections: Inhibiting inositol lipid signaling by targeting Sec14 phosphatidylinositol transfer proteins.Adv Biol Regul. 2022 May;84:100891. doi: 10.1016/j.jbior.2022.100891. Epub 2022 Feb 25. Adv Biol Regul. 2022. PMID: 35240534 Free PMC article. Review.
References
-
- Heitman J, Kozel TR, Kwon-Chung KJ, Perfect JR, Casadevall A. 2010. Cryptococcus: from human pathogen to model yeast, 1st ed, vol 1, p 620 American Society for Microbiology, Washington, DC
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

