Fine tuning of the catalytic activity of colicin E7 nuclease domain by systematic N-terminal mutations
- PMID: 24895333
- PMCID: PMC4116659
- DOI: 10.1002/pro.2497
Fine tuning of the catalytic activity of colicin E7 nuclease domain by systematic N-terminal mutations
Abstract
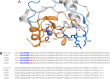
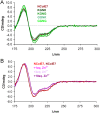
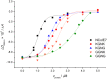
The nuclease domain of colicin E7 (NColE7) promotes the nonspecific cleavage of nucleic acids at its C-terminal HNH motif. Interestingly, the deletion of four N-terminal residues (446-449 NColE7 = KRNK) resulted in complete loss of the enzyme activity. R447A mutation was reported to decrease the nuclease activity, but a detailed analysis of the role of the highly positive and flexible N-terminus is still missing. Here, we present the study of four mutants, with a decreased activity in the following order: NColE7 >> KGNK > KGNG ∼ GGNK > GGNG. At the same time, the folding, the metal-ion, and the DNA-binding affinity were unaffected by the mutations as revealed by linear and circular dichroism spectroscopy, isothermal calorimetric titrations, and gel mobility shift experiments. Semiempirical quantum chemical calculations and molecular dynamics simulations revealed that K446, K449, and/or the N-terminal amino group are able to approach the active centre in the absence of the other positively charged residues. The results suggested a complex role of the N-terminus in the catalytic process that could be exploited in the design of a controlled nuclease.
Keywords: DNA cleavage; Zn2+; binding; flow linear dichroism; isothermal calorimetry; positively charged N-terminal residues.
© 2014 The Protein Society.
Figures




References
-
- Chak K, Kuo W, Lu F, James R. Cloning and characterization of the Cole7 plasmid. J Gen Microbiol. 1991;137:91–100. - PubMed
-
- Lin Y, Liao C, Liang P, Yuan H, Chak K. Involvement of colicin in the limited protection of the colicin producing cells against bacteriophage. Biochem Biophys Res Commun. 2004;318:81–87. - PubMed
-
- Liao C, Hsiao K, Liu Y, Leng P, Yuen HS, Chak K. Processing of DNase domain during translocation of colicin E7 across the membrane of Escherichia coli. Biochem Biophys Res Commun. 2001;284:556–562. - PubMed
-
- Cheng YS, Shi Z, Doudeva LG, Yang WZ, Chak KF, Yuan HS. High-resolution crystal structure of a truncated ColE7 translocation domain: implications for colicin transport across membranes. J Mol Biol. 2006;356:22–31. - PubMed
-
- Shi Z, Chak K, Yuan H. Identification of an essential cleavage site in ColE7 required for import and killing of cells. J Biol Chem. 2005;280:24663–24668. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

