DiseaseConnect: a comprehensive web server for mechanism-based disease-disease connections
- PMID: 24895436
- PMCID: PMC4086092
- DOI: 10.1093/nar/gku412
DiseaseConnect: a comprehensive web server for mechanism-based disease-disease connections
Abstract
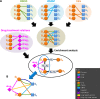
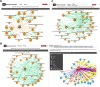
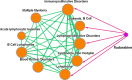
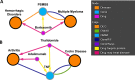
The DiseaseConnect (http://disease-connect.org) is a web server for analysis and visualization of a comprehensive knowledge on mechanism-based disease connectivity. The traditional disease classification system groups diseases with similar clinical symptoms and phenotypic traits. Thus, diseases with entirely different pathologies could be grouped together, leading to a similar treatment design. Such problems could be avoided if diseases were classified based on their molecular mechanisms. Connecting diseases with similar pathological mechanisms could inspire novel strategies on the effective repositioning of existing drugs and therapies. Although there have been several studies attempting to generate disease connectivity networks, they have not yet utilized the enormous and rapidly growing public repositories of disease-related omics data and literature, two primary resources capable of providing insights into disease connections at an unprecedented level of detail. Our DiseaseConnect, the first public web server, integrates comprehensive omics and literature data, including a large amount of gene expression data, Genome-Wide Association Studies catalog, and text-mined knowledge, to discover disease-disease connectivity via common molecular mechanisms. Moreover, the clinical comorbidity data and a comprehensive compilation of known drug-disease relationships are additionally utilized for advancing the understanding of the disease landscape and for facilitating the mechanism-based development of new drug treatments.
© The Author(s) 2014. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures
Similar articles
-
Infertility etiologies are genetically and clinically linked with other diseases in single meta-diseases.Reprod Biol Endocrinol. 2015 Apr 15;13:31. doi: 10.1186/s12958-015-0029-9. Reprod Biol Endocrinol. 2015. PMID: 25880215 Free PMC article. Review.
-
Mergeomics 2.0: a web server for multi-omics data integration to elucidate disease networks and predict therapeutics.Nucleic Acids Res. 2021 Jul 2;49(W1):W375-W387. doi: 10.1093/nar/gkab405. Nucleic Acids Res. 2021. PMID: 34048577 Free PMC article.
-
Mergeomics: a web server for identifying pathological pathways, networks, and key regulators via multidimensional data integration.BMC Genomics. 2016 Sep 9;17(1):722. doi: 10.1186/s12864-016-3057-8. BMC Genomics. 2016. PMID: 27612452 Free PMC article.
-
DIANA-microT Web server upgrade supports Fly and Worm miRNA target prediction and bibliographic miRNA to disease association.Nucleic Acids Res. 2011 Jul;39(Web Server issue):W145-8. doi: 10.1093/nar/gkr294. Epub 2011 May 6. Nucleic Acids Res. 2011. PMID: 21551220 Free PMC article.
-
A compilation of Web-based research tools for miRNA analysis.Brief Funct Genomics. 2017 Sep 1;16(5):249-273. doi: 10.1093/bfgp/elw042. Brief Funct Genomics. 2017. PMID: 28334134 Review.
Cited by
-
Combined network pharmacology and virtual reverse pharmacology approaches for identification of potential targets to treat vascular dementia.Sci Rep. 2020 Jan 14;10(1):257. doi: 10.1038/s41598-019-57199-9. Sci Rep. 2020. PMID: 31937840 Free PMC article.
-
A network analysis of cofactor-protein interactions for analyzing associations between human nutrition and diseases.Sci Rep. 2016 Jan 18;6:19633. doi: 10.1038/srep19633. Sci Rep. 2016. PMID: 26777674 Free PMC article.
-
Applications of network analysis to routinely collected health care data: a systematic review.J Am Med Inform Assoc. 2018 Feb 1;25(2):210-221. doi: 10.1093/jamia/ocx052. J Am Med Inform Assoc. 2018. PMID: 29025116 Free PMC article.
-
A small molecule targeting CHI3L1 inhibits lung metastasis by blocking IL-13Rα2-mediated JNK-AP-1 signals.Mol Oncol. 2022 Jan;16(2):508-526. doi: 10.1002/1878-0261.13138. Epub 2021 Nov 24. Mol Oncol. 2022. PMID: 34758182 Free PMC article.
-
A Systems Approach to Refine Disease Taxonomy by Integrating Phenotypic and Molecular Networks.EBioMedicine. 2018 May;31:79-91. doi: 10.1016/j.ebiom.2018.04.002. Epub 2018 Apr 6. EBioMedicine. 2018. PMID: 29669699 Free PMC article.
References
-
- Van Driel M.A., Bruggeman J., Vriend G., Brunner H.G., Leunissen J.A.M. A text-mining analysis of the human phenome. Eur. J. Hum. Genet. 2006;14:535–542. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

