Construction and characterization of a bacterial artificial chromosome library for the hexaploid wheat line 92R137
- PMID: 24895618
- PMCID: PMC4026951
- DOI: 10.1155/2014/845806
Construction and characterization of a bacterial artificial chromosome library for the hexaploid wheat line 92R137
Abstract
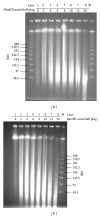
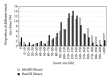
For map-based cloning of genes conferring important traits in the hexaploid wheat line 92R137, a bacterial artificial chromosome (BAC) library, including two sublibraries, was constructed using the genomic DNA of 92R137 digested with restriction enzymes HindIII and BamHI. The BAC library was composed of total 765,696 clones, of which 390,144 were from the HindIII digestion and 375,552 from the BamHI digestion. Through pulsed-field gel electrophoresis (PFGE) analysis of 453 clones randomly selected from the HindIII sublibrary and 573 clones from the BamHI sublibrary, the average insert sizes were estimated as 129 and 113 kb, respectively. Thus, the HindIII sublibrary was estimated to have a 3.01-fold coverage and the BamHI sublibrary a 2.53-fold coverage based on the estimated hexaploid wheat genome size of 16,700 Mb. The 765,696 clones were arrayed in 1,994 384-well plates. All clones were also arranged into plate pools and further arranged into 5-dimensional (5D) pools. The probability of identifying a clone corresponding to any wheat DNA sequence (such as gene Yr26 for stripe rust resistance) from the library was estimated to be more than 99.6%. Through polymerase chain reaction screening the 5D pools with Xwe173, a marker tightly linked to Yr26, six BAC clones were successfully obtained. These results demonstrate that the BAC library is a valuable genomic resource for positional cloning of Yr26 and other genes of interest.
Figures


Similar articles
-
Construction and characterization of three wheat bacterial artificial chromosome libraries.Int J Mol Sci. 2014 Nov 28;15(12):21896-912. doi: 10.3390/ijms151221896. Int J Mol Sci. 2014. PMID: 25464379 Free PMC article.
-
Construction of a hexaploid wheat (Triticum aestivum L.) bacterial artificial chromosome library for cloning genes for stripe rust resistance.Genome. 2005 Dec;48(6):1028-36. doi: 10.1139/g05-078. Genome. 2005. PMID: 16391672
-
Construction and characterization of a bacterial artificial chromosome (BAC) library of hexaploid wheat (Triticum aestivum L.) and validation of genome coverage using locus-specific primers.Genome. 2003 Oct;46(5):870-8. doi: 10.1139/g03-067. Genome. 2003. PMID: 14608404
-
Construction of a subgenomic BAC library specific for chromosomes 1D, 4D and 6D of hexaploid wheat.Theor Appl Genet. 2004 Nov;109(7):1337-45. doi: 10.1007/s00122-004-1768-8. Epub 2004 Sep 10. Theor Appl Genet. 2004. PMID: 15365624
-
Rapid identification of the three homoeologues of the wheat dwarfing gene Rht using a novel PCR-based screen of three-dimensional BAC pools.Genome. 2009 Dec;52(12):993-1000. doi: 10.1139/G09-073. Genome. 2009. PMID: 19953127
Cited by
-
Construction and characterization of three wheat bacterial artificial chromosome libraries.Int J Mol Sci. 2014 Nov 28;15(12):21896-912. doi: 10.3390/ijms151221896. Int J Mol Sci. 2014. PMID: 25464379 Free PMC article.
-
β-mercaptoethanol assists efficient construction of sperm bacterial artificial chromosome library.J Biol Methods. 2017 Jan 20;4(1):e63. doi: 10.14440/jbm.2017.167. eCollection 2017. J Biol Methods. 2017. PMID: 31453223 Free PMC article.
-
Haplotype-resolved telomere-to-telomere genome assembly of the dikaryotic fungus pathogen Rhizoctonia cerealis.Sci Data. 2025 Jun 6;12(1):951. doi: 10.1038/s41597-025-05260-w. Sci Data. 2025. PMID: 40481055 Free PMC article.
-
Genome-wide identification of the Q-type C2H2 zinc finger protein gene family and expression analysis under abiotic stress in lotus (Nelumbo nucifera G.).BMC Genomics. 2024 Jun 28;25(1):648. doi: 10.1186/s12864-024-10546-1. BMC Genomics. 2024. PMID: 38943098 Free PMC article.
References
-
- Huang X-Q, Röder MS. Molecular mapping of powdery mildew resistance genes in wheat: a review. Euphytica. 2004;137(2):203–223.
-
- Sharma-Poudyal D, Chen X, Wan A, et al. Virulence characterization of international collections of the wheat stripe rust pathogen, Puccinia striiformis f. sp. tritici . Plant Disease. 2013;97(3):379–386. - PubMed
-
- Chen XM. Epidemiology and control of stripe rust [Puccinia striiformis f. sp. tritici] on wheat. Canadian Journal of Plant Pathology. 2005;27(3):314–337.
-
- Chen WQ, Wu LR, Liu TG, et al. Race dynamics, diversity, and virulence evolution in Puccinia striiformis f. sp. tritici, the causal agent of wheat stripe rust in china from 2003 to 2007. Plant Disease. 2009;93(11):1093–1101. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

