Fidelity of cotranslational protein targeting by the signal recognition particle
- PMID: 24895856
- PMCID: PMC4444370
- DOI: 10.1146/annurev-biophys-051013-022653
Fidelity of cotranslational protein targeting by the signal recognition particle
Abstract
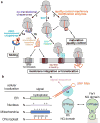
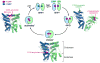
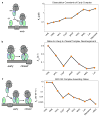
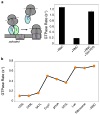
Accurate folding, assembly, localization, and maturation of newly synthesized proteins are essential to all cells and require high fidelity in the protein biogenesis machineries that mediate these processes. Here, we review our current understanding of how high fidelity is achieved in one of these processes, the cotranslational targeting of nascent membrane and secretory proteins by the signal recognition particle (SRP). Recent biochemical, biophysical, and structural studies have elucidated how the correct substrates drive a series of elaborate conformational rearrangements in the SRP and SRP receptor GTPases; these rearrangements provide effective fidelity checkpoints to reject incorrect substrates and enhance the fidelity of this essential cellular pathway. The mechanisms used by SRP to ensure fidelity share important conceptual analogies with those used by cellular machineries involved in DNA replication, transcription, and translation, and these mechanisms likely represent general principles for other complex cellular pathways.
Keywords: GTPases; RNA; protein biogenesis; protein translocation; ribosome; signal sequence.
Figures


References
-
- Albanese V, Yam AY-W, Baughman J, Parnot C, Frydman J. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell. 2006;124:75–88. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

