miR-148a- and miR-216a-regulated oncolytic adenoviruses targeting pancreatic tumors attenuate tissue damage without perturbation of miRNA activity
- PMID: 24895996
- PMCID: PMC4435498
- DOI: 10.1038/mt.2014.98
miR-148a- and miR-216a-regulated oncolytic adenoviruses targeting pancreatic tumors attenuate tissue damage without perturbation of miRNA activity
Abstract
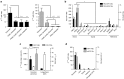
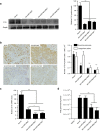
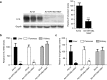
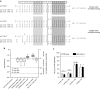
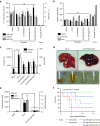
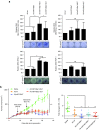
Oncolytic virotherapy shows promise for pancreatic ductal adenocarcinoma (PDAC) treatment, but there is the need to minimize associated-toxicities. In the current work, we engineered artificial target sites recognized by miR-216a and/or miR-148a to provide pancreatic tumor-selectivity to replication-competent adenoviruses (Ad-miRTs) and improve their safety profile. Expression analysis in PDAC patients identified miR-148a and miR-216a downregulated in resectable (FC(miR-148a) = 0.044, P < 0.05; FC(miR-216a) = 0.017, P < 0.05), locally advanced (FC(miR-148a) = 0.038, P < 0.001; FC(miR-216a) = 0.001, P < 0.001) and metastatic tumors (FC(miR-148a) = 0.041, P < 0.01; FC(miR-216a) = 0.002, P < 0.001). In mouse tissues, miR-216a was highly specific of the exocrine pancreas whereas miR-148a was abundant in the exocrine pancreas, Langerhans islets, and the liver. In line with the miRNA content and the miRNA target site design, we show E1A gene expression and viral propagation efficiently controlled in Ad-miRT-infected cells. Consequently, Ad-miRT-infected mice presented reduced pancreatic and liver damage without perturbation of the endogenous miRNAs and their targets. Interestingly, the 8-miR148aT design showed repressing activity by all miR-148/152 family members with significant detargeting effects in the pancreas and liver. Ad-miRTs preserved their oncolytic activity and triggered strong antitumoral responses. This study provides preclinical evidences of miR-148a and miR-216a target site insertions to confer adenoviral selectivity and proposes 8-miR148aT as an optimal detargeting strategy for genetically-engineered therapies against PDAC.
Figures

References
-
- Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol Rev. 2011;91:827–887. - PubMed
-
- Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007;297:1901–1908. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

