Improved imputation quality of low-frequency and rare variants in European samples using the 'Genome of The Netherlands'
- PMID: 24896149
- PMCID: PMC4200431
- DOI: 10.1038/ejhg.2014.19
Improved imputation quality of low-frequency and rare variants in European samples using the 'Genome of The Netherlands'
Abstract
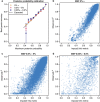
Although genome-wide association studies (GWAS) have identified many common variants associated with complex traits, low-frequency and rare variants have not been interrogated in a comprehensive manner. Imputation from dense reference panels, such as the 1000 Genomes Project (1000G), enables testing of ungenotyped variants for association. Here we present the results of imputation using a large, new population-specific panel: the Genome of The Netherlands (GoNL). We benchmarked the performance of the 1000G and GoNL reference sets by comparing imputation genotypes with 'true' genotypes typed on ImmunoChip in three European populations (Dutch, British, and Italian). GoNL showed significant improvement in the imputation quality for rare variants (MAF 0.05-0.5%) compared with 1000G. In Dutch samples, the mean observed Pearson correlation, r(2), increased from 0.61 to 0.71. We also saw improved imputation accuracy for other European populations (in the British samples, r(2) improved from 0.58 to 0.65, and in the Italians from 0.43 to 0.47). A combined reference set comprising 1000G and GoNL improved the imputation of rare variants even further. The Italian samples benefitted the most from this combined reference (the mean r(2) increased from 0.47 to 0.50). We conclude that the creation of a large population-specific reference is advantageous for imputing rare variants and that a combined reference panel across multiple populations yields the best imputation results.
Figures

References
-
- Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

