A MLVA genotyping scheme for global surveillance of the citrus pathogen Xanthomonas citri pv. citri suggests a worldwide geographical expansion of a single genetic lineage
- PMID: 24897119
- PMCID: PMC4045669
- DOI: 10.1371/journal.pone.0098129
A MLVA genotyping scheme for global surveillance of the citrus pathogen Xanthomonas citri pv. citri suggests a worldwide geographical expansion of a single genetic lineage
Abstract
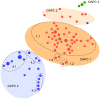
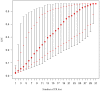
MultiLocus Variable number of tandem repeat Analysis (MLVA) has been extensively used to examine epidemiological and evolutionary issues on monomorphic human pathogenic bacteria, but not on bacterial plant pathogens of agricultural importance albeit such tools would improve our understanding of their epidemiology, as well as of the history of epidemics on a global scale. Xanthomonas citri pv. citri is a quarantine organism in several countries and a major threat for the citrus industry worldwide. We screened the genomes of Xanthomonas citri pv. citri strain IAPAR 306 and of phylogenetically related xanthomonads for tandem repeats. From these in silico data, an optimized MLVA scheme was developed to assess the global diversity of this monomorphic bacterium. Thirty-one minisatellite loci (MLVA-31) were selected to assess the genetic structure of 129 strains representative of the worldwide pathological and genetic diversity of X. citri pv. citri. Based on Discriminant Analysis of Principal Components (DAPC), four pathotype-specific clusters were defined. DAPC cluster 1 comprised strains that were implicated in the major geographical expansion of X. citri pv. citri during the 20th century. A subset of 12 loci (MLVA-12) resolved 89% of the total diversity and matched the genetic structure revealed by MLVA-31. MLVA-12 is proposed for routine epidemiological identification of X. citri pv. citri, whereas MLVA-31 is proposed for phylogenetic and population genetics studies. MLVA-31 represents an opportunity for international X. citri pv. citri genotyping and data sharing. The MLVA-31 data generated in this study was deposited in the Xanthomonas citri genotyping database (http://www.biopred.net/MLVA/).
Conflict of interest statement
Figures
Similar articles
-
From local surveys to global surveillance: three high-throughput genotyping methods for epidemiological monitoring of Xanthomonas citri pv. citri pathotypes.Appl Environ Microbiol. 2009 Feb;75(4):1173-84. doi: 10.1128/AEM.02245-08. Epub 2008 Dec 16. Appl Environ Microbiol. 2009. PMID: 19088309 Free PMC article.
-
Core-Genome Multilocus Sequence Typing for Epidemiological and Evolutionary Analyses of Phytopathogenic Xanthomonas citri.Appl Environ Microbiol. 2023 May 31;89(5):e0210122. doi: 10.1128/aem.02101-22. Epub 2023 Apr 17. Appl Environ Microbiol. 2023. PMID: 37067413 Free PMC article.
-
Amplified fragment length polymorphism and multilocus sequence analysis-based genotypic relatedness among pathogenic variants of Xanthomonas citri pv. citri and Xanthomonas campestris pv. bilvae.Int J Syst Evol Microbiol. 2010 Mar;60(Pt 3):515-525. doi: 10.1099/ijs.0.009514-0. Epub 2009 Aug 4. Int J Syst Evol Microbiol. 2010. PMID: 19654364
-
Highly polymorphic markers reveal the establishment of an invasive lineage of the citrus bacterial pathogen Xanthomonas citri pv. citri in its area of origin.Environ Microbiol. 2014 Jul;16(7):2226-37. doi: 10.1111/1462-2920.12369. Epub 2014 Jan 21. Environ Microbiol. 2014. PMID: 24373118
-
Genetic and structural determinants on iron assimilation pathways in the plant pathogen Xanthomonas citri subsp. citri and Xanthomonas sp.Braz J Microbiol. 2020 Sep;51(3):1219-1231. doi: 10.1007/s42770-019-00207-x. Epub 2020 Aug 28. Braz J Microbiol. 2020. PMID: 31848911 Free PMC article. Review.
Cited by
-
Origin and diversification of Xanthomonas citri subsp. citri pathotypes revealed by inclusive phylogenomic, dating, and biogeographic analyses.BMC Genomics. 2019 Sep 9;20(1):700. doi: 10.1186/s12864-019-6007-4. BMC Genomics. 2019. PMID: 31500575 Free PMC article.
-
Development of a Multiple Loci Variable Number of Tandem Repeats Analysis (MLVA) to Unravel the Intra-Pathovar Structure of Pseudomonas syringae pv. actinidiae Populations Worldwide.PLoS One. 2015 Aug 11;10(8):e0135310. doi: 10.1371/journal.pone.0135310. eCollection 2015. PLoS One. 2015. PMID: 26262683 Free PMC article.
-
Novel Plastid-Nuclear Genome Combinations Enhance Resistance to Citrus Canker in Cybrid Grapefruit.Front Plant Sci. 2019 Jan 7;9:1858. doi: 10.3389/fpls.2018.01858. eCollection 2018. Front Plant Sci. 2019. PMID: 30666259 Free PMC article.
-
Contrasting genetic diversity and structure among Malagasy Ralstonia pseudosolanacearum phylotype I populations inferred from an optimized Multilocus Variable Number of Tandem Repeat Analysis scheme.PLoS One. 2020 Dec 8;15(12):e0242846. doi: 10.1371/journal.pone.0242846. eCollection 2020. PLoS One. 2020. PMID: 33290390 Free PMC article.
-
Development and comparative validation of genomic-driven PCR-based assays to detect Xanthomonas citri pv. citri in citrus plants.BMC Microbiol. 2020 Oct 1;20(1):296. doi: 10.1186/s12866-020-01972-8. BMC Microbiol. 2020. PMID: 33004016 Free PMC article.
References
-
- Urwin R, Maiden MCJ (2003) Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol 11: 479–487. - PubMed
-
- Maiden MCJ (2006) Multilocus sequence typing of bacteria. Annual Review of Microbiology 60: 561–588. - PubMed
-
- Achtman M (2008) Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu Rev Microbiol 62: 53–70. - PubMed
-
- Kado CI (2010) Plant bacteriology. Saint Paul, MN, USA: The American Phytopathological Society.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

