Selenoprotein S is involved in maintenance and transport of multiprotein complexes
- PMID: 24897171
- PMCID: PMC5578454
- DOI: 10.1042/BJ20140076
Selenoprotein S is involved in maintenance and transport of multiprotein complexes
Abstract
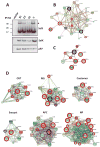
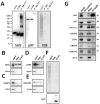
SelS (Selenoprotein S) is a selenocysteine-containing protein with roles in ER (endoplasmic reticulum) function and inflammation. It has been implicated in ERAD (ER-associated protein degradation), and clinical studies revealed an association of its promoter polymorphism with cytokine levels and human diseases. However, the pathways and interacting proteins that could shed light on pathogenesis of SelS-associated diseases have not been studied systematically. We performed a large-scale affinity isolation of human SelS and its mutant forms and analysed the proteins that interact with them. All previously known SelS targets and nearly two hundred additional proteins were identified that were remarkably enriched for various multiprotein complexes. Subsequent chemical cross-linking experiments identified the specific interacting sites in SelS and its several targets. Most of these interactions involved coiled-coil domains. The data suggest that SelS participates in intracellular membrane transport and maintenance of protein complexes by anchoring them to the ER membrane.
Figures



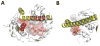
References
-
- Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. - PubMed
-
- McCann JCA, Bruce N. Adaptive dysfunction of selenoproteins from the perspective of the triage theory: why modest selenium deficiency may increase risk of diseases of aging. FASEB Journal. 2011;25:1793–1814. - PubMed
-
- Papp LV, Holmgren A, Khanna KK. Selenium and selenoproteins in health and disease. Antioxid Redox Signal. 2010;12:793–795. - PubMed
-
- Ye Y, Shibata Y, Yun C, Ron D, Rapoport TA. A membrane protein complex mediates retro-translocation from the ER lumen into the cytosol. Nature. 2004;429:841–847. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

