Comprehensive secondary structure elucidation of four genera of the family Pospiviroidae
- PMID: 24897295
- PMCID: PMC4045682
- DOI: 10.1371/journal.pone.0098655
Comprehensive secondary structure elucidation of four genera of the family Pospiviroidae
Abstract
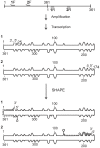
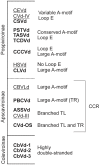
Viroids are small, circular, single stranded RNA molecules that infect plants. Since they are non-coding, their structures play a critical role in their life cycles. To date, little effort has been spend on elucidating viroid structures in solution due to both the experimental difficulties and the time-consuming nature of the methodologies implicated. Recently, the technique of high-throughput selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE) was adapted for the probing of the members of family Avsunviroidae, all of whom replicate in the chloroplast and demonstrate ribozyme activity. In the present work, twelve viroid species belonging to four different genera of the family Pospiviroidae, whose members are characterized by the presence of a central conserved region (CCR) and who replicate in nucleus of the host, were probed. Given that the structures of five distinct viroid species from the family Pospiviroidae have been previously reported, an overview of the different structural characteristics for all genera and the beginning of a manual classification of the different viroids based on their structural features are presented here.
Conflict of interest statement
Figures



References
-
- Ding B (2010) Viroids: Self-replicating, mobile, and fast-evolving noncoding regulatory RNAs. Wiley Interdiscip Rev RNA 1: 362–375. - PubMed
-
- Diener TO (2003) Discovering viroids—a personal perspective. Nat Rev Microbiol 1: 75–80. - PubMed
-
- Flores R, Hernandez C, Martinez de Alba AE, Daros JA, Di Serio F (2005) Viroids and viroid-host interactions. Annu Rev Phytopathol 43: 117–139. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

