Genes found essential in other mycoplasmas are dispensable in Mycoplasma bovis
- PMID: 24897538
- PMCID: PMC4045577
- DOI: 10.1371/journal.pone.0097100
Genes found essential in other mycoplasmas are dispensable in Mycoplasma bovis
Abstract
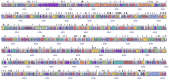
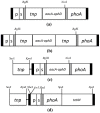
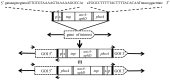
Mycoplasmas are regarded to be useful models for studying the minimum genetic complement required for independent survival of an organism. Mycoplasma bovis is a globally distributed pathogen causing pneumonia, mastitis, arthritis, otitis media and reproductive tract disease, and genome sequences of three strains, the type strain PG45 and two strains isolated in China, have been published. In this study, several Tn4001 based transposon constructs were generated and used to create a M. bovis PG45 insertional mutant library. Direct genome sequencing of 319 independent insertions detected disruptions in 129 genes in M. bovis, 48 of which had homologues in Mycoplasma mycoides subspecies mycoides SC and 99 of which had homologues in Mycoplasma agalactiae. Sixteen genes found to be essential in previous studies on other mycoplasma species were found to be dispensable. Five of these genes have previously been predicted to be part of the core set of 153 essential genes in mycoplasmas. Thus this study has extended the list of non-essential genes of mycoplasmas from that previously generated by studies in other species.
Conflict of interest statement
Figures
References
-
- Caswell JL, Archambault M (2007) Mycoplasma bovis pneumonia in cattle. Animal Health Research Reviews 8: 161–186. - PubMed
-
- Dybvig K, Voelker LL (1996) Molecular biology of mycoplasmas. Annual Review of Microbiology 50: 25–57. - PubMed
-
- Halbedel S, Stulke J (2007) Tools for the genetic analysis of Mycoplasma. International Journal of Medical Microbiology 297: 37–44. - PubMed
-
- Minion FC (2002) Molecular pathogenesis of mycoplasma animal respiratory pathogens. Frontiers in Bioscience 7: d1410–1422. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

