Detection of an en masse and reversible B- to A-DNA conformational transition in prokaryotes in response to desiccation
- PMID: 24898023
- PMCID: PMC4208382
- DOI: 10.1098/rsif.2014.0454
Detection of an en masse and reversible B- to A-DNA conformational transition in prokaryotes in response to desiccation
Abstract
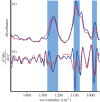
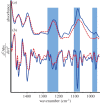
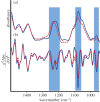
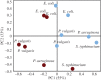
The role that DNA conformation plays in the biochemistry of cells has been the subject of intensive research since DNA polymorphism was discovered. B-DNA has long been considered the native form of DNA in cells although alternative conformations of DNA are thought to occur transiently and along short tracts. Here, we report the first direct observation of a fully reversible en masse conformational transition between B- and A-DNA within live bacterial cells using Fourier transform infrared (FTIR) spectroscopy. This biospectroscopic technique allows for non-invasive and reagent-free examination of the holistic biochemistry of samples. For this reason, we have been able to observe the previously unknown conformational transition in all four species of bacteria investigated. Detection of this transition is evidence of a previously unexplored biological significance for A-DNA and highlights the need for new research into the role that A-DNA plays as a cellular defence mechanism and in stabilizing the DNA conformation. Such studies are pivotal in understanding the role of A-DNA in the evolutionary pathway of nucleic acids. Furthermore, this discovery demonstrates the exquisite capabilities of FTIR spectroscopy and opens the door for further investigations of cell biochemistry with this under-used technique.
Keywords: A-DNA; DNA conformation; attenuated total reflection–Fourier transform infrared spectroscopy; bacteria; infrared.
Figures
Similar articles
-
Monitoring the reversible B to A-like transition of DNA in eukaryotic cells using Fourier transform infrared spectroscopy.Nucleic Acids Res. 2011 Jul;39(13):5439-48. doi: 10.1093/nar/gkr175. Epub 2011 Mar 29. Nucleic Acids Res. 2011. PMID: 21447564 Free PMC article.
-
Understanding B-DNA to A-DNA transition in the right-handed DNA helix: Perspective from a local to global transition.Prog Biophys Mol Biol. 2017 Sep;128:63-73. doi: 10.1016/j.pbiomolbio.2017.05.009. Epub 2017 May 30. Prog Biophys Mol Biol. 2017. PMID: 28576665 Review.
-
Concerted dynamics of metallo-base pairs in an A/B-form helical transition.Nat Commun. 2019 Oct 23;10(1):4818. doi: 10.1038/s41467-019-12440-x. Nat Commun. 2019. PMID: 31645548 Free PMC article.
-
Free-Energy Profiles for A-/B-DNA Conformational Transitions in Isolated and Aggregated States from All-Atom Molecular Dynamics Simulation.J Phys Chem B. 2018 Aug 23;122(33):7990-7996. doi: 10.1021/acs.jpcb.8b04573. Epub 2018 Aug 14. J Phys Chem B. 2018. PMID: 30067905
-
The importance of hydration and DNA conformation in interpreting infrared spectra of cells and tissues.Chem Soc Rev. 2016 Apr 7;45(7):1980-98. doi: 10.1039/c5cs00511f. Chem Soc Rev. 2016. PMID: 26403652 Review.
Cited by
-
Transitions of Double-Stranded DNA Between the A- and B-Forms.J Phys Chem B. 2016 Aug 25;120(33):8449-56. doi: 10.1021/acs.jpcb.6b02155. Epub 2016 May 11. J Phys Chem B. 2016. PMID: 27135262 Free PMC article.
-
Kinetics of the Conformational Transformation between B- and A-Forms in the Drew-Dickerson Dodecamer.ACS Omega. 2020 Dec 15;5(51):32995-33006. doi: 10.1021/acsomega.0c04247. eCollection 2020 Dec 29. ACS Omega. 2020. PMID: 33403261 Free PMC article.
-
Quantum Mechanics Characterization of Non-Covalent Interaction in Nucleotide Fragments.Molecules. 2024 Jul 10;29(14):3258. doi: 10.3390/molecules29143258. Molecules. 2024. PMID: 39064837 Free PMC article.
-
A survey of recent unusual high-resolution DNA structures provoked by mismatches, repeats and ligand binding.Nucleic Acids Res. 2018 Jul 27;46(13):6416-6434. doi: 10.1093/nar/gky561. Nucleic Acids Res. 2018. PMID: 29945186 Free PMC article. Review.
-
In vivo atomic force microscopy-infrared spectroscopy of bacteria.J R Soc Interface. 2018 Mar;15(140):20180115. doi: 10.1098/rsif.2018.0115. J R Soc Interface. 2018. PMID: 29593091 Free PMC article.
References
-
- Crick FHC, Watson JD. 1954. The complementary structure of deoxyribonucleic acid. Proc. R. Soc. Lond. A 223, 80–96. (10.1098/rspa.1954.0101) - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

