Limited dissemination and shedding of the UL128 complex-intact, UL/b'-defective rhesus cytomegalovirus strain 180.92
- PMID: 24899204
- PMCID: PMC4136270
- DOI: 10.1128/JVI.00162-14
Limited dissemination and shedding of the UL128 complex-intact, UL/b'-defective rhesus cytomegalovirus strain 180.92
Abstract
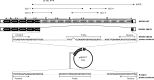
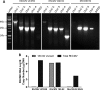
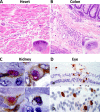
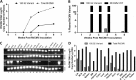
The UL128 complex of human cytomegalovirus (CMV) is a major determinant of viral entry into epithelial and endothelial cells and a target for vaccine development. The UL/b' region of rhesus CMV contains several open reading frames, including orthologs of the UL128 complex. We recently showed that the coding content of the rhesus CMV (RhCMV) UL/b' region predicts acute endothelial tropism and long-term shedding in vivo in the rhesus macaque model of CMV infection. The laboratory-passaged RhCMV 180.92 strain has a truncated UL/b' region but an intact UL128 complex. To investigate whether the presence of the UL128 complex alone was sufficient to confer endothelial and epithelial tropism in vivo, we investigated tissue dissemination and viral excretion following experimental RhCMV 180.92 inoculation of RhCMV-seronegative rhesus macaques. We show the presence of at least two virus variants in the RhCMV 180.92 infectious virus stock. A rare variant noted for a nontruncated wild-type-virus-like UL/b' region, rapidly emerged during in vivo replication and showed high-level replication in blood and tissues and excretion in urine and saliva, features similar to those previously reported in naturally occurring wild-type RhCMV infection. In contrast, the predominant truncated version of RhCMV 180.92 showed significantly lower plasma DNAemia and limited tissue dissemination and viral shedding. These data demonstrate that the truncated RhCMV 180.92 variant is attenuated in vivo and suggest that additional UL/b' genes, besides the UL128 complex, are required for optimal in vivo CMV replication and dissemination.
Importance: An effective vaccine against human CMV infection will need to target genes that are essential for virus propagation and transmission. The human CMV UL128 complex represents one such candidate antigen since it is essential for endothelial and epithelial cell tropism, and is a target for neutralizing antibodies in CMV-infected individuals. In this study, we used the rhesus macaque animal model of CMV infection to investigate the in vivo function of the UL128 complex. Using experimental infection of rhesus macaques with a rhesus CMV virus variant that contained an intact UL128 complex but was missing several other genes, we show that the presence of the UL128 complex alone is not sufficient for widespread tissue dissemination and virus excretion. These data highlight the importance of in vivo studies in evaluating human CMV gene function and suggest that additional UL/b' genes are required for optimal CMV dissemination and transmission.
Copyright © 2014, American Society for Microbiology. All Rights Reserved.
Figures
Similar articles
-
Open reading frames carried on UL/b' are implicated in shedding and horizontal transmission of rhesus cytomegalovirus in rhesus monkeys.J Virol. 2011 May;85(10):5105-14. doi: 10.1128/JVI.02631-10. Epub 2011 Mar 9. J Virol. 2011. PMID: 21389128 Free PMC article.
-
The susceptibility of primary cultured rhesus macaque kidney epithelial cells to rhesus cytomegalovirus strains.J Gen Virol. 2016 Jun;97(6):1426-1438. doi: 10.1099/jgv.0.000455. Epub 2016 Mar 14. J Gen Virol. 2016. PMID: 26974598 Free PMC article.
-
Exploitation of Interleukin-10 (IL-10) Signaling Pathways: Alternate Roles of Viral and Cellular IL-10 in Rhesus Cytomegalovirus Infection.J Virol. 2016 Oct 14;90(21):9920-9930. doi: 10.1128/JVI.00635-16. Print 2016 Nov 1. J Virol. 2016. PMID: 27558431 Free PMC article.
-
The power of human cytomegalovirus (HCMV) hijacked UL/b' functions lost in vitro.Acta Virol. 2020;64(2):117-130. doi: 10.4149/av_2020_202. Acta Virol. 2020. PMID: 32551781 Review.
-
Characteristics and functions of human cytomegalovirus UL128 gene/protein.Acta Virol. 2014;58(2):103-7. doi: 10.4149/av_2014_02_103. Acta Virol. 2014. PMID: 24957713 Review.
Cited by
-
Protective effect of pre-existing natural immunity in a nonhuman primate reinfection model of congenital cytomegalovirus infection.PLoS Pathog. 2023 Oct 5;19(10):e1011646. doi: 10.1371/journal.ppat.1011646. eCollection 2023 Oct. PLoS Pathog. 2023. PMID: 37796819 Free PMC article.
-
Protective effect of pre-existing natural immunity in a nonhuman primate reinfection model of congenital cytomegalovirus infection.bioRxiv [Preprint]. 2023 Apr 10:2023.04.10.536057. doi: 10.1101/2023.04.10.536057. bioRxiv. 2023. Update in: PLoS Pathog. 2023 Oct 5;19(10):e1011646. doi: 10.1371/journal.ppat.1011646. PMID: 37090643 Free PMC article. Updated. Preprint.
-
Cytomegalovirus Vaccines: Current Status and Future Prospects.Drugs. 2016 Nov;76(17):1625-1645. doi: 10.1007/s40265-016-0653-5. Drugs. 2016. PMID: 27882457 Free PMC article. Review.
-
In vitro and in vivo characterization of a recombinant rhesus cytomegalovirus containing a complete genome.PLoS Pathog. 2020 Nov 24;16(11):e1008666. doi: 10.1371/journal.ppat.1008666. eCollection 2020 Nov. PLoS Pathog. 2020. PMID: 33232376 Free PMC article.
-
Impact of CMV Infection on Natural Killer Cell Clonal Repertoire in CMV-Naïve Rhesus Macaques.Front Immunol. 2019 Oct 9;10:2381. doi: 10.3389/fimmu.2019.02381. eCollection 2019. Front Immunol. 2019. PMID: 31649681 Free PMC article.
References
-
- Fields BN, Knipe DM, Howley PM. 2007. Fields virology, 5th ed. Wolters Kluwer Health/Lippincott/The Williams & Wilkins Co, Philadelphia, PA
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

