Evolutionary insights into premetazoan functions of the neuronal protein homer
- PMID: 24899667
- PMCID: PMC4137706
- DOI: 10.1093/molbev/msu178
Evolutionary insights into premetazoan functions of the neuronal protein homer
Abstract
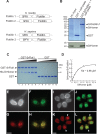
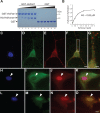
Reconstructing the evolution and ancestral functions of synaptic proteins promises to shed light on how neurons first evolved. The postsynaptic density (PSD) protein Homer scaffolds membrane receptors and regulates Ca(2+) signaling in diverse metazoan cell types (including neurons and muscle cells), yet its ancestry and core functions are poorly understood. We find that the protein domain organization and essential biochemical properties of metazoan Homer proteins, including their ability to tetramerize, are conserved in the choanoflagellate Salpingoeca rosetta, one of the closest living relatives of metazoans. Unlike in neurons, Homer localizes to the nucleoplasm in S. rosetta and interacts directly with Flotillin, a protein more commonly associated with cell membranes. Surprisingly, we found that the Homer/Flotillin interaction and its localization to the nucleus are conserved in metazoan astrocytes. These findings suggest that Homer originally interacted with Flotillin in the nucleus of the last common ancestor of metazoans and choanoflagellates and was later co-opted to function as a membrane receptor scaffold in the PSD.
Keywords: Flotillin; Homer; S. rosetta; choanoflagellate; postsynaptic scaffold evolution; synapse.
© The Author 2014. Published by Oxford University Press on behalf of the Society for Molecular Biology and Evolution.
Figures



References
-
- Baldauf SL. The deep roots of eukaryotes. Science. 2003;300:1703–1706. - PubMed
-
- Bickel PE, Scherer PE, Schnitzer JE, Oh P, Lisanti MP, Lodish HF. Flotillin and epidermal surface antigen define a new family of caveolae-associated integral membrane proteins. J Biol Chem. 1997;272:13793–13802. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

