Genetically targeted binary labeling of retinal neurons
- PMID: 24899708
- PMCID: PMC4044247
- DOI: 10.1523/JNEUROSCI.2960-13.2014
Genetically targeted binary labeling of retinal neurons
Abstract
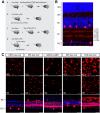
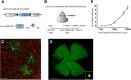
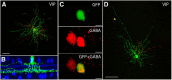
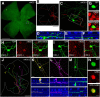
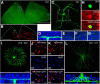
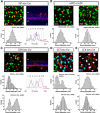
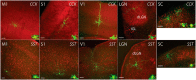
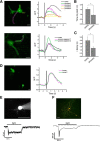
A major stumbling block to understanding neural circuits is the extreme anatomical and functional diversity of interneurons. Subsets of interneurons can be targeted for manipulation using Cre mouse lines, but Cre expression is rarely confined to a single interneuron type. It is essential to have a strategy that further restricts labeling in Cre driver lines. We now describe an approach that combines Cre driver mice, recombinant adeno-associated virus, and rabies virus to produce sparse but binary labeling of select interneurons--frequently only a single cell in a large region. We used this approach to characterize the retinal amacrine and ganglion cell types in five GABAergic Cre mouse (Mus musculus) lines, and identified two new amacrine cell types: an asymmetric medium-field type and a wide-field type. We also labeled several wide-field amacrine cell types that have been previously identified based on morphology but whose connectivity and function had not been systematically studied due to lack of genetic markers. All Cre-expressing amacrine cells labeled with an antibody to GABA. Cre-expressing RGCs lacked GABA labeling and included classically defined as well as recently identified types. In addition to the retina, our technique leads to sparse labeling of neurons in the cortex, lateral geniculate nucleus, and superior colliculus, and can be used to express optogenetic tools such as channelrhodopsin and protein sensors such as GCaMP. The Cre drivers identified in this study provide genetic access to otherwise hard to access cell types for systematic analysis including anatomical characterization, physiological recording, optogenetic and/or chemical manipulation, and circuit mapping.
Keywords: Cre recombinase; amacrine cell; ganglion cell; rabies virus; retina.
Copyright © 2014 the authors 0270-6474/14/347845-17$15.00/0.
Figures




Similar articles
-
Heterogeneous transgene expression in the retinas of the TH-RFP, TH-Cre, TH-BAC-Cre and DAT-Cre mouse lines.Neuroscience. 2015 Oct 29;307:319-37. doi: 10.1016/j.neuroscience.2015.08.060. Epub 2015 Aug 31. Neuroscience. 2015. PMID: 26335381 Free PMC article.
-
Characterization of green fluorescent protein-expressing retinal cells in CD 44-transgenic mice.Neuroscience. 2007 Feb 9;144(3):1087-93. doi: 10.1016/j.neuroscience.2006.09.061. Epub 2006 Dec 8. Neuroscience. 2007. PMID: 17161542 Free PMC article.
-
Age-related alterations in neurons of the mouse retina.J Neurosci. 2011 Nov 2;31(44):16033-44. doi: 10.1523/JNEUROSCI.3580-11.2011. J Neurosci. 2011. PMID: 22049445 Free PMC article.
-
Cre Activated and Inactivated Recombinant Adeno-Associated Viral Vectors for Neuronal Anatomical Tracing or Activity Manipulation.Curr Protoc Neurosci. 2015 Jul 1;72:1.24.1-1.24.15. doi: 10.1002/0471142301.ns0124s72. Curr Protoc Neurosci. 2015. PMID: 26131660 Free PMC article. Review.
-
Genetic dissection of GABAergic neural circuits in mouse neocortex.Front Cell Neurosci. 2014 Jan 27;8:8. doi: 10.3389/fncel.2014.00008. eCollection 2014. Front Cell Neurosci. 2014. PMID: 24478631 Free PMC article. Review.
Cited by
-
nGnG Amacrine Cells and Brn3b-negative M1 ipRGCs are Specifically Labeled in the ChAT-ChR2-EYFP Mouse.Invest Ophthalmol Vis Sci. 2020 Feb 7;61(2):14. doi: 10.1167/iovs.61.2.14. Invest Ophthalmol Vis Sci. 2020. PMID: 32049344 Free PMC article.
-
Identification of retinal ganglion cell types and brain nuclei expressing the transcription factor Brn3c/Pou4f3 using a Cre recombinase knock-in allele.J Comp Neurol. 2021 Jun;529(8):1926-1953. doi: 10.1002/cne.25065. Epub 2020 Nov 10. J Comp Neurol. 2021. PMID: 33135183 Free PMC article.
-
Functionally distinct GABAergic amacrine cell types regulate spatiotemporal encoding in the mouse retina.Nat Neurosci. 2025 Jun;28(6):1256-1267. doi: 10.1038/s41593-025-01935-0. Epub 2025 Apr 15. Nat Neurosci. 2025. PMID: 40234708 Free PMC article.
-
Morphology and function of three VIP-expressing amacrine cell types in the mouse retina.J Neurophysiol. 2015 Oct;114(4):2431-8. doi: 10.1152/jn.00526.2015. Epub 2015 Aug 26. J Neurophysiol. 2015. PMID: 26311183 Free PMC article.
-
The CRF Family of Neuropeptides and their Receptors - Mediators of the Central Stress Response.Curr Mol Pharmacol. 2018;11(1):4-31. doi: 10.2174/1874467210666170302104053. Curr Mol Pharmacol. 2018. PMID: 28260504 Free PMC article. Review.
References
-
- Beier KT, Saunders A, Oldenburg IA, Miyamichi K, Akhtar N, Luo L, Whelan SP, Sabatini B, Cepko CL. Anterograde or retrograde transsynaptic labeling of CNS neurons with vesicular stomatitis virus vectors. Proc Natl Acad Sci U S A. 2011;108:15414–15419. doi: 10.1073/pnas.1110854108. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
