Discovery of Pyridyl Bis(oxy)dibenzimidamide Derivatives as Selective Matriptase Inhibitors
- PMID: 24900621
- PMCID: PMC4027570
- DOI: 10.1021/ml400213v
Discovery of Pyridyl Bis(oxy)dibenzimidamide Derivatives as Selective Matriptase Inhibitors
Abstract
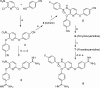
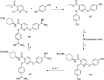
Matriptase belongs to trypsin-like serine proteases involved in matrix remodeling/degradation, growth regulation, survival, motility, and cell morphogenesis. Herein, we report a structure-based approach, which led to the discovery of sulfonamide and amide derivatives of pyridyl bis(oxy)benzamidine as potent and selective matriptase inhibitors. Co-crystal structures of selected compounds in complex with matriptase supported compound designing. Additionally, WaterMap analyses indicated the possibility of occupying a distinct pocket within the catalytic domain, exploration of which resulted in >100-fold improvement in potency. Co-crystal structure of 10 with matriptase revealed critical interactions leading to potent target inhibition and selectivity against other serine proteases.
Keywords: Matriptase; SAR; cancer; crystal structure; pyridyl dibenzimidamide.
Figures


References
-
- Coghlin C.; Murray G. I. Current and emerging concepts in tumour metastasis. J. Pathol. 2010, 222, 1–15. - PubMed
-
- Duffy M. J. Do proteases play a role in cancer invasion and metastasis?. Eur. J. Cancer Clin. Oncol. 1987, 23, 583–589. - PubMed
-
- Liotta L. A.; Stetler-Stevenson W. G. Tumor invasion and metastasis: an imbalance of positive and negative regulation. Cancer Res. 1991, 51, 5054s–5059s. - PubMed
-
- Deryugina E. I.; Quigley J. P. Matrix metalloproteinases and tumor metastasis. Cancer Metastasis Rev. 2006, 25, 9–34. - PubMed
-
- Bonfil R. D.; Cher M. L. The role of proteolytic enzymes in metastatic bone disease. IBMS BoneKEy 2011, 8, 16–36.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information
Miscellaneous

