Fragment-based discovery of 6-azaindazoles as inhibitors of bacterial DNA ligase
- PMID: 24900632
- PMCID: PMC4027552
- DOI: 10.1021/ml4003277
Fragment-based discovery of 6-azaindazoles as inhibitors of bacterial DNA ligase
Abstract
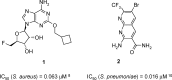
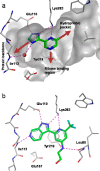
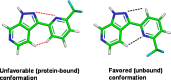
Herein we describe the application of fragment-based drug design to bacterial DNA ligase. X-ray crystallography was used to guide structure-based optimization of a fragment-screening hit to give novel, nanomolar, AMP-competitive inhibitors. The lead compound 13 showed antibacterial activity across a range of pathogens. Data to demonstrate mode of action was provided using a strain of S. aureus, engineered to overexpress DNA ligase.
Keywords: Bacterial DNA ligase; S. aureus; fragment-based drug design; structure-based optimization.
Figures

Similar articles
-
Novel bacterial NAD+-dependent DNA ligase inhibitors with broad-spectrum activity and antibacterial efficacy in vivo.Antimicrob Agents Chemother. 2011 Mar;55(3):1088-96. doi: 10.1128/AAC.01181-10. Epub 2010 Dec 28. Antimicrob Agents Chemother. 2011. PMID: 21189350 Free PMC article.
-
Pyrrolamide DNA gyrase inhibitors: fragment-based nuclear magnetic resonance screening to identify antibacterial agents.Antimicrob Agents Chemother. 2012 Mar;56(3):1240-6. doi: 10.1128/AAC.05485-11. Epub 2011 Dec 19. Antimicrob Agents Chemother. 2012. PMID: 22183167 Free PMC article.
-
Fragment-to-hit-to-lead discovery of a novel pyridylurea scaffold of ATP competitive dual targeting type II topoisomerase inhibiting antibacterial agents.J Med Chem. 2013 Nov 14;56(21):8712-35. doi: 10.1021/jm401208b. Epub 2013 Oct 29. J Med Chem. 2013. PMID: 24098982
-
Structure guided design of biotin protein ligase inhibitors for antibiotic discovery.Curr Top Med Chem. 2014;14(1):4-20. doi: 10.2174/1568026613666131111103149. Curr Top Med Chem. 2014. PMID: 24236729 Review.
-
Combining NMR and X-ray crystallography in fragment-based drug discovery: discovery of highly potent and selective BACE-1 inhibitors.Top Curr Chem. 2012;317:83-114. doi: 10.1007/128_2011_183. Top Curr Chem. 2012. PMID: 21647837 Review.
Cited by
-
Detection of secondary binding sites in proteins using fragment screening.Proc Natl Acad Sci U S A. 2015 Dec 29;112(52):15910-5. doi: 10.1073/pnas.1518946112. Epub 2015 Dec 11. Proc Natl Acad Sci U S A. 2015. PMID: 26655740 Free PMC article.
-
Synthesis and Antiproliferative Activity of 2,4,6,7-Tetrasubstituted-2H-pyrazolo[4,3-c]pyridines.Molecules. 2021 Nov 8;26(21):6747. doi: 10.3390/molecules26216747. Molecules. 2021. PMID: 34771163 Free PMC article.
-
Targeting DNA Replication and Repair for the Development of Novel Therapeutics against Tuberculosis.Front Mol Biosci. 2017 Nov 14;4:75. doi: 10.3389/fmolb.2017.00075. eCollection 2017. Front Mol Biosci. 2017. PMID: 29184888 Free PMC article. Review.
-
The Macromolecular Machines that Duplicate the Escherichia coli Chromosome as Targets for Drug Discovery.Antibiotics (Basel). 2018 Mar 14;7(1):23. doi: 10.3390/antibiotics7010023. Antibiotics (Basel). 2018. PMID: 29538288 Free PMC article. Review.
-
Compound Uptake into E. coli Can Be Facilitated by N-Alkyl Guanidiniums and Pyridiniums.ACS Infect Dis. 2021 Jan 8;7(1):162-173. doi: 10.1021/acsinfecdis.0c00715. Epub 2020 Nov 23. ACS Infect Dis. 2021. PMID: 33228356 Free PMC article.
References
-
- Lehman I. R. DNA ligase: Structure, mechanism and function. Science 1974, 186, 790. - PubMed
-
- Tomkinson A. E.; Vijayakumar S.; Pascal J. M.; Ellenberger T. DNA ligase: Structure, reaction mechanism and function. Chem. Rev. 2006, 106, 687. - PubMed
-
- Lavesa-Curto M.; Sayer H.; Bullard D.; MacDonald A.; Wilkinson A.; Smith A.; Bowater L.; Hemmings A.; Bowater R. P. Characterisation of a temperature-sensitive DNA ligase from Escherichia coli. Microbiology 2004, 150, 4171–4180. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Chemical Information

