Deep evolutionary comparison of gene expression identifies parallel recruitment of trans-factors in two independent origins of C4 photosynthesis
- PMID: 24901697
- PMCID: PMC4046924
- DOI: 10.1371/journal.pgen.1004365
Deep evolutionary comparison of gene expression identifies parallel recruitment of trans-factors in two independent origins of C4 photosynthesis
Erratum in
-
Correction: Deep Evolutionary Comparison of Gene Expression Identifies Parallel Recruitment of Trans-Factors in Two Independent Origins of C4 Photosynthesis.PLoS Genet. 2016 May 24;12(5):e1006087. doi: 10.1371/journal.pgen.1006087. eCollection 2016 May. PLoS Genet. 2016. PMID: 27220009 Free PMC article.
Abstract
With at least 60 independent origins spanning monocotyledons and dicotyledons, the C4 photosynthetic pathway represents one of the most remarkable examples of convergent evolution. The recurrent evolution of this highly complex trait involving alterations to leaf anatomy, cell biology and biochemistry allows an increase in productivity by ∼ 50% in tropical and subtropical areas. The extent to which separate lineages of C4 plants use the same genetic networks to maintain C4 photosynthesis is unknown. We developed a new informatics framework to enable deep evolutionary comparison of gene expression in species lacking reference genomes. We exploited this to compare gene expression in species representing two independent C4 lineages (Cleome gynandra and Zea mays) whose last common ancestor diverged ∼ 140 million years ago. We define a cohort of 3,335 genes that represent conserved components of leaf and photosynthetic development in these species. Furthermore, we show that genes encoding proteins of the C4 cycle are recruited into networks defined by photosynthesis-related genes. Despite the wide evolutionary separation and independent origins of the C4 phenotype, we report that these species use homologous transcription factors to both induce C4 photosynthesis and to maintain the cell specific gene expression required for the pathway to operate. We define a core molecular signature associated with leaf and photosynthetic maturation that is likely shared by angiosperm species derived from the last common ancestor of the monocotyledons and dicotyledons. We show that deep evolutionary comparisons of gene expression can reveal novel insight into the molecular convergence of highly complex phenotypes and that parallel evolution of trans-factors underpins the repeated appearance of C4 photosynthesis. Thus, exploitation of extant natural variation associated with complex traits can be used to identify regulators. Moreover, the transcription factors that are shared by independent C4 lineages are key targets for engineering the C4 pathway into C3 crops such as rice.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
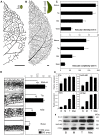
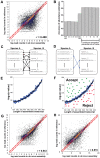
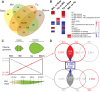

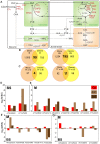
References
-
- Christin PA, Osborne CP, Sage RF, Arakaki M, Edwards EJ (2011) C4 eudicots are not younger than C4 monocots. J Exp Bot 62: 3171–3181. - PubMed
-
- Sage RF, Christin PA, Edwards EJ (2011) The C4 plant lineages of planet Earth. J Exp Bot 62: 3155–3169. - PubMed
-
- Brown HA (1999) Agronomic implications of C4 photosynthesis. In C4 Plant Biology. In: (Sage RFaM, R.K.,eds). San Diego, CA: Academic Press, editor. pp. 473–508.
-
- Hatch MD (1987) C4 photosynthesis: a unique elend of modified biochemistry, anatomy and ultrastructure. Biochimica et Biophysica Acta (BBA) - Reviews on Bioenergetics 895: 81–106.
-
- Hibberd JM, Covshoff S (2010) The Regulation of Gene Expression Required for C4 Photosynthesis. Annu Rev Plant Biol 61: 181–207. - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

