C3HC4-type RING finger protein NbZFP1 is involved in growth and fruit development in Nicotiana benthamiana
- PMID: 24901716
- PMCID: PMC4047095
- DOI: 10.1371/journal.pone.0099352
C3HC4-type RING finger protein NbZFP1 is involved in growth and fruit development in Nicotiana benthamiana
Abstract
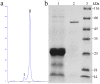
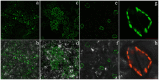
C3HC4-type RING finger proteins constitute a large family in the plant kingdom and play important roles in various physiological processes of plant life. In this study, a C3HC4-type zinc finger gene was isolated from Nicotiana benthamiana. Sequence analysis indicated that the gene encodes a 24-kDa protein with 191 amino acids containing one typical C3HC4-type zinc finger domain; this gene was named NbZFP1. Transient expression of pGDG-NbZFP1 demonstrated that NbZFP1 was localized to the chloroplast, especially in the chloroplasts of cells surrounding leaf stomata. Virus-induced gene silencing (VIGS) analysis indicated that silencing of NbZFP1 hampered fruit development, although the height of the plants was normal. An overexpression construct was then designed and transferred into Nicotiana benthamiana, and PCR and Southern blot showed that the NbZFP1 gene was successfully integrated into the Nicotiana benthamiana genome. The transgenic lines showed typical compactness, with a short internode length and sturdy stems. This is the first report describing the function of a C3HC4-type RING finger protein in tobacco.
Conflict of interest statement
Figures




References
-
- Berg JM, Shi Y (1996) The galvanization of biology: a growing appreciation for the roles of zinc. Science 271: 1081–1085. - PubMed
-
- Von Arnim AG, Deng X (1993) Ring finger motif of Arabidopsis thaliana COP1 defines a new class of zinc-binding domain. Journal of Biological Chemistry 268: 19626–19631. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

