The ubiquitin-conjugating enzyme, UbcM2, is restricted to monoubiquitylation by a two-fold mechanism that involves backside residues of E2 and Lys48 of ubiquitin
- PMID: 24901938
- PMCID: PMC4072368
- DOI: 10.1021/bi500072v
The ubiquitin-conjugating enzyme, UbcM2, is restricted to monoubiquitylation by a two-fold mechanism that involves backside residues of E2 and Lys48 of ubiquitin
Abstract
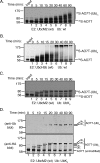
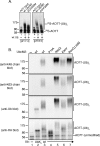
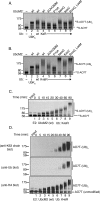
Proteins can be modified on lysines (K) with a single ubiquitin (Ub) or with polymers of Ub (polyUb). These different configurations and their respective topologies are primary factors for determining whether substrates are targeted to the proteasome for degradation or directed to nonproteolytic outcomes. We report here on the intrinsic ubiquitylation properties of UbcM2 (UBE2E3/UbcH9), a conserved Ub-conjugating enzyme linked to cell proliferation, development, and the cellular antioxidant defense system. Using a fully recombinant ubiquitylation assay, we show that UbcM2 is severely limited in its ability to synthesize polyUb chains with wild-type Ub. Restriction to monoubiquitylation is governed by multiple residues on the backside of the enzyme, far removed from its active site, and by lysine 48 of Ub. UbcM2 with mutated backside residues can synthesize K63-linked polyUb chains and to a lesser extent K6- and K48-linked chains. Additionally, we identified a single residue on the backside of the enzyme that promotes monoubiquitylation. Together, these findings reveal that a combination of noncatalytic residues within the Ubc catalytic core domain of UbcM2 as well as a lysine(s) within Ub can relegate a Ub-conjugating enzyme to monoubiquitylate its cognate targets despite having the latent capacity to construct polyUb chains. The two-fold mechanism for restricting activity to monoubiquitylation provides added insurance that UbcM2 will not build polyUb chains on its substrates, even under conditions of high local Ub concentrations.
Figures




References
-
- Chau V.; Tobias J. W.; Bachmair A.; Marriott D.; Ecker D. J.; Gonda D. K.; Varshavsky A. (1989) A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science 243, 1576–1583. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

