Salmonella Typhi and Salmonella Paratyphi A elaborate distinct systemic metabolite signatures during enteric fever
- PMID: 24902583
- PMCID: PMC4077204
- DOI: 10.7554/eLife.03100
Salmonella Typhi and Salmonella Paratyphi A elaborate distinct systemic metabolite signatures during enteric fever
Abstract
The host-pathogen interactions induced by Salmonella Typhi and Salmonella Paratyphi A during enteric fever are poorly understood. This knowledge gap, and the human restricted nature of these bacteria, limit our understanding of the disease and impede the development of new diagnostic approaches. To investigate metabolite signals associated with enteric fever we performed two dimensional gas chromatography with time-of-flight mass spectrometry (GCxGC/TOFMS) on plasma from patients with S. Typhi and S. Paratyphi A infections and asymptomatic controls, identifying 695 individual metabolite peaks. Applying supervised pattern recognition, we found highly significant and reproducible metabolite profiles separating S. Typhi cases, S. Paratyphi A cases, and controls, calculating that a combination of six metabolites could accurately define the etiological agent. For the first time we show that reproducible and serovar specific systemic biomarkers can be detected during enteric fever. Our work defines several biologically plausible metabolites that can be used to detect enteric fever, and unlocks the potential of this method in diagnosing other systemic bacterial infections.
Keywords: Salmonella Paratyphi A; Salmonella Typhi; enteric fever; epidemiology; global health; human; infectious disease; mass spectrometry; metabolites; microbiology; typhoid.
Conflict of interest statement
The authors declare that no competing interests exist.
Figures

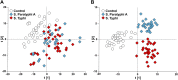


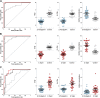

Comment in
-
Honing in on enteric fever.Elife. 2014 Jul 1;3:e03545. doi: 10.7554/eLife.03545. Elife. 2014. PMID: 24986861 Free PMC article.
References
-
- Arjyal A, Basnyat B, Koirala S, Karkey A, Dongol S, Agrawaal KK, Shakya N, Shrestha K, Sharma M, Lama S, Shrestha K, Khatri NS, Shrestha U, Campbell JI, Baker S, Farrar J, Wolbers M, Dolecek C. 2011. Gatifloxacin versus chloramphenicol for uncomplicated enteric fever: an open-label, randomised, controlled trial. The Lancet Infectious Diseases 11:445–454. doi: 10.1016/S1473-3099(11)70089-5 - DOI - PMC - PubMed
-
- Baker S, Holt KE, Clements AC, Karkey A, Arjyal A, Boni MF, Dongol S, Hammond N, Koirala S, Duy PT, Nga TV, Campbell JI, Dolecek C, Basnyat B, Dougan G, Farrar JJ. 2011. Combined high-resolution genotyping and geospatial analysis reveals modes of endemic urban typhoid fever transmission. Open Biology 1:110008. doi: 10.1098/rsob.110008 - DOI - PMC - PubMed
-
- Baumgarner BL, Cooper BR. 2012. Evaluation of a tandem gas chromatography/time-of-flight mass spectrometry metabolomics platform as a single method to investigate the effect of starvation on whole-animal metabolism in rainbow trout (Oncorhynchus mykiss). The Journal of Experimental Biology 215:1627–1632 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

